DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis
- PMID: 25023518
- PMCID: PMC4107794
- DOI: 10.1083/jcb.201311063
DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis
Abstract
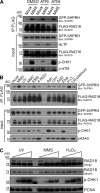
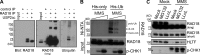
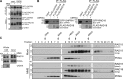
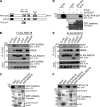
Deoxyribonucleic acid (DNA) lesions encountered during replication are often bypassed using DNA damage tolerance (DDT) pathways to avoid prolonged fork stalling and allow for completion of DNA replication. Rad18 is a central E3 ubiquitin ligase in DDT, which exists in a monoubiquitinated (Rad18•Ub) and nonubiquitinated form in human cells. We find that Rad18 is deubiquitinated when cells are treated with methyl methanesulfonate or hydrogen peroxide. The ubiquitinated form of Rad18 does not interact with SNF2 histone linker plant homeodomain RING helicase (SHPRH) or helicase-like transcription factor, two downstream E3 ligases needed to carry out error-free bypass of DNA lesions. Instead, it interacts preferentially with the zinc finger domain of another, nonubiquitinated Rad18 and may inhibit Rad18 function in trans. Ubiquitination also prevents Rad18 from localizing to sites of DNA damage, inducing proliferating cell nuclear antigen monoubiquitination, and suppressing mutagenesis. These data reveal a new role for monoubiquitination in controlling Rad18 function and suggest that damage-specific deubiquitination promotes a switch from Rad18•Ub-Rad18 complexes to the Rad18-SHPRH complexes necessary for error-free lesion bypass in cells.
© 2014 Zeman et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

