T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity
- PMID: 25024161
- PMCID: PMC4199372
- DOI: 10.1101/gr.170753.113
T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity
Abstract
The T-cell receptor (TCR) repertoire is formed by random recombinations of genomic precursor elements; the resulting combinatorial diversity renders unlikely extensive TCR sharing between individuals. Here, we studied CDR3β amino acid sequence sharing in a repertoire-wide manner, using high-throughput TCR-seq in 28 healthy mice. We uncovered hundreds of public sequences shared by most mice. Public CDR3 sequences, relative to private sequences, are two orders of magnitude more abundant on average, express restricted V/J segments, and feature high convergent nucleic acid recombination. Functionally, public sequences are enriched for MHC-diverse CDR3 sequences that were previously associated with autoimmune, allograft, and tumor-related reactions, but not with anti-pathogen-related reactions. Public CDR3 sequences are shared between mice of different MHC haplotypes, but are associated with different, MHC-dependent, V genes. Thus, despite their random generation process, TCR repertoires express a degree of uniformity in their post-genomic organization. These results, together with numerical simulations of TCR genomic rearrangements, suggest that biases and convergence in TCR recombination combine with ongoing selection to generate a restricted subset of self-associated, public CDR3 TCR sequences, and invite reexamination of the basic mechanisms of T-cell repertoire formation.
© 2014 Madi et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
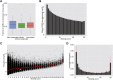


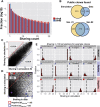
 ). (Blue region) 1.15% of the sequences show much higher sharing in the data (
). (Blue region) 1.15% of the sequences show much higher sharing in the data ( ). (E) Measured (vertical dashed line) and simulated (red bars, histograms of 100 random runs of simulation A) sharing levels of four selected CDR3 aa sequences from each region in D. (Top) Neutral, showing similar sharing in simulations and data (purple circles in D,
). (E) Measured (vertical dashed line) and simulated (red bars, histograms of 100 random runs of simulation A) sharing levels of four selected CDR3 aa sequences from each region in D. (Top) Neutral, showing similar sharing in simulations and data (purple circles in D,  ); (middle) positive, showing much higher sharing in data vs. simulation (blue triangles in D,
); (middle) positive, showing much higher sharing in data vs. simulation (blue triangles in D,  ); (bottom) negative (red squares in D,
); (bottom) negative (red squares in D,  ) showing much lower sharing in the data. Two sequences in each row are taken from the annotated group of sequences shown in Figure 4.
) showing much lower sharing in the data. Two sequences in each row are taken from the annotated group of sequences shown in Figure 4.
References
-
- Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. 1998. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity 9: 169–178 - PubMed
-
- Burnet FM. 1976. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J Clin 26: 119–121 - PubMed
-
- Casrouge A, Beaudoing E, Dalle S, Pannetier C, Kanellopoulos J, Kourilsky P. 2000. Size estimate of the αβ TCR repertoire of naive mouse splenocytes. J Immunol 164: 5782–5787 - PubMed
-
- Cohen IR. 1992. The cognitive principle challenges clonal selection. Immunol Today 13: 441–444 - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
