Spatial interactions and cooperation can change the speed of evolution of complex phenotypes
- PMID: 25024187
- PMCID: PMC4113926
- DOI: 10.1073/pnas.1400828111
Spatial interactions and cooperation can change the speed of evolution of complex phenotypes
Abstract
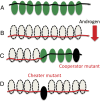
Complex traits arise from the interactions among multiple gene products. In the case where the complex phenotype is separated from the wild type by a fitness valley or a fitness plateau, the generation of a complex phenotype may take a very long evolutionary time. Interestingly, the rate of evolution depends in nontrivial ways on various properties of the underlying stochastic process, such as the spatial organization of the population and social interactions among cells. Here we review some of our recent work that investigates these phenomena in asexual populations. The role of spatial constraints is quite complex: there are realistic cases where spatial constrains can accelerate or delay evolution, or even influence it in a nonmonotonic fashion, where evolution works fastest for intermediate-range constraints. Social interactions among cells can be studied in the context of the division-of-labor games. Under a range of circumstances, cooperation among cells can lead to a relatively fast creation of a complex phenotype as an emerging (distributed) property. If we further assume the presence of cheaters, we observe the emergence of a fully mutated population of cells possessing the complex phenotype. Applications of these ideas to cancer initiation and biofilm formation in bacteria are discussed.
Keywords: mutations; stochastic modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–2048. - PubMed
-
- Serra MC, Haccou P. Dynamics of escape mutants. Theor Popul Biol. 2007;72(1):167–178. - PubMed
-
- Weinreich DM, Chao L. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution. 2005;59(6):1175–1182. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

