Arhgap36-dependent activation of Gli transcription factors
- PMID: 25024229
- PMCID: PMC4121843
- DOI: 10.1073/pnas.1322362111
Arhgap36-dependent activation of Gli transcription factors
Abstract
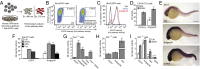
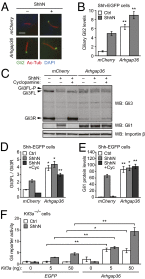
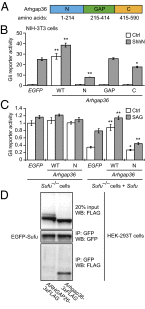
Hedgehog (Hh) pathway activation and Gli-dependent transcription play critical roles in embryonic patterning, tissue homeostasis, and tumorigenesis. By conducting a genome-scale cDNA overexpression screen, we have identified the Rho GAP family member Arhgap36 as a positive regulator of the Hh pathway in vitro and in vivo. Arhgap36 acts in a Smoothened (Smo)-independent manner to inhibit Gli repressor formation and to promote the activation of full-length Gli proteins. Arhgap36 concurrently induces the accumulation of Gli proteins in the primary cilium, and its ability to induce Gli-dependent transcription requires kinesin family member 3a and intraflagellar transport protein 88, proteins that are essential for ciliogenesis. Arhgap36 also functionally and biochemically interacts with Suppressor of Fused. Transcriptional profiling further reveals that Arhgap36 is overexpressed in murine medulloblastomas that acquire resistance to chemical Smo inhibitors and that ARHGAP36 isoforms capable of Gli activation are up-regulated in a subset of human medulloblastomas. Our findings reveal a new mechanism of Gli transcription factor activation and implicate ARHGAP36 dysregulation in the onset and/or progression of GLI-dependent cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

