OMICtools: an informative directory for multi-omic data analysis
- PMID: 25024350
- PMCID: PMC4095679
- DOI: 10.1093/database/bau069
OMICtools: an informative directory for multi-omic data analysis
Abstract
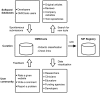
Recent advances in 'omic' technologies have created unprecedented opportunities for biological research, but current software and database resources are extremely fragmented. OMICtools is a manually curated metadatabase that provides an overview of more than 4400 web-accessible tools related to genomics, transcriptomics, proteomics and metabolomics. All tools have been classified by omic technologies (next-generation sequencing, microarray, mass spectrometry and nuclear magnetic resonance) associated with published evaluations of tool performance. Information about each tool is derived either from a diverse set of developers, the scientific literature or from spontaneous submissions. OMICtools is expected to serve as a useful didactic resource not only for bioinformaticians but also for experimental researchers and clinicians. Database URL: http://omictools.com/.
© The Author(s) 2014. Published by Oxford University Press.
Figures

References
-
- Metzker M.L. (2010) Sequencing technologies—the next generation. Nat. Rev. Genet., 11, 31–46 - PubMed
-
- Hoheisel J.D. (2006) Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev. Genet., 7, 200–210 - PubMed
-
- Nilsson T., Mann M., Aebersold R., et al. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods, 7, 681–685 - PubMed
-
- Nicholson J.K., Lindon J.C. (2008) Systems biology: metabonomics. Nature, 455, 1054–1056 - PubMed
-
- Nekrutenko A., Taylor J. (2012) Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet., 13, 667–672 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

