A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress
- PMID: 25024436
- PMCID: PMC4195785
- DOI: 10.15252/embj.201488566
A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress
Abstract
Living cells compartmentalize materials and enzymatic reactions to increase metabolic efficiency. While eukaryotes use membrane-bound organelles, bacteria and archaea rely primarily on protein-bound nanocompartments. Encapsulins constitute a class of nanocompartments widespread in bacteria and archaea whose functions have hitherto been unclear. Here, we characterize the encapsulin nanocompartment from Myxococcus xanthus, which consists of a shell protein (EncA, 32.5 kDa) and three internal proteins (EncB, 17 kDa; EncC, 13 kDa; EncD, 11 kDa). Using cryo-electron microscopy, we determined that EncA self-assembles into an icosahedral shell 32 nm in diameter (26 nm internal diameter), built from 180 subunits with the fold first observed in bacteriophage HK97 capsid. The internal proteins, of which EncB and EncC have ferritin-like domains, attach to its inner surface. Native nanocompartments have dense iron-rich cores. Functionally, they resemble ferritins, cage-like iron storage proteins, but with a massively greater capacity (~30,000 iron atoms versus ~3,000 in ferritin). Physiological data reveal that few nanocompartments are assembled during vegetative growth, but they increase fivefold upon starvation, protecting cells from oxidative stress through iron sequestration.
Keywords: HK97 fold; cryo‐electron microscopy; encapsulin; ferritin; oxidative stress.
© 2014 The Authors.
Figures

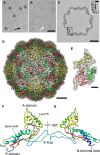
EM of negatively stained nanocompartments. Scale bar, 100 nm.
SDS–PAGE of purifed native encapsulin nanocompartments. Mass markers are indicated (in kDa).
Domain maps of the four encapsulin proteins. Yellow, HK97-like domain; orange, rubrerythrin/ferritin-like domain; asterisks, ExxH metal coordination motifs; gray, encapsulation signal.
EM of unstained/air-dried nanocompartments. Scale bar, 100 nm.
STEM and energy-dispersive X-ray spectroscopy (EDX) images of two representative particles. In the STEM image (unstained specimen), positive signals from mass scattering identify two nanocompartments, while the corresponding EDX images map elemental concentrations of Fe and P (strongly above background), and S and Ca (at or close to background). Red crosshairs indicate identical positions on one nanocompartment. Scale bar, 100 nm.
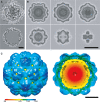
Cryo-EM image of a native encapsulin nanocompartment without (top) and with (bottom) masking out of the electron-dense core.
Sections through the single-particle reconstruction of the native Myxococcus xanthus encapsulin nanocompartment.
Isosurface representation of the reconstruction color-coded according to radial distance from the center (given in Å). Left, viewed from the outside; right, cutaway view of the internal structure.
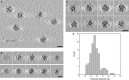
Tomographic slice showing several native Myxococcus xanthus encapsulin nanocompartments.
Tomographic slices through two nanocompartments (top and bottom rows).
Gallery of tomographic central sections of eight nanocompartments.
Histogram of the diameters of electron-dense granules in the nanocompartment cores.



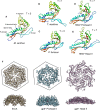

Shown are cutaway views of the EncA shell (blue) for the particles with (left) and without (right) iron-rich granules. The C-terminal anchor motif common to EncB, EncC, and EncD is depicted as a small gray oval. In EncB and EncC, this motif is connected by an extended flexible linker to the rubrerythrin domain (green ovals). EncD's domain is shown as a brown oval. Since EncB and EncC are predicted to have only two α-helices each, we hypothesize that they dimerize forming a 4-helix bundle in homo- or hetero-dimers with the iron-binding motifs midway along the bundle. However, monomers may also exist, and they may be capable of nucleating iron deposition. The model envisages iron atoms entering the shell through narrow channels, to be incorporated into a nascent granule nucleated on the rubrerythrin domains. Interaction of the iron with the internal proteins probably induces a conformational change that causes the rubrerythrin domains to detach from the EncA shell.
The most likely candidates for the entry channels are shown.
References
-
- Abrescia NG, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. - PubMed
-
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. The Phenix refinement framework. CCP4 Newsl 42, contribution 8.
-
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage T7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15:461–472. - PubMed
-
- Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, Nakaishi Y, Suzuki M, Namba K, Ono Y, Tsukihara T, Nakagawa A. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol. 2007;368:1469–1483. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

