Single nucleotide polymorphisms with cis-regulatory effects on long non-coding transcripts in human primary monocytes
- PMID: 25025429
- PMCID: PMC4099216
- DOI: 10.1371/journal.pone.0102612
Single nucleotide polymorphisms with cis-regulatory effects on long non-coding transcripts in human primary monocytes
Abstract
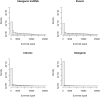
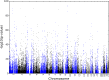
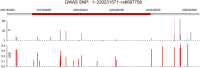
We applied genome-wide allele-specific expression analysis of monocytes from 188 samples. Monocytes were purified from white blood cells of healthy blood donors to detect cis-acting genetic variation that regulates the expression of long non-coding RNAs. We analysed 8929 regions harboring genes for potential long non-coding RNA that were retrieved from data from the ENCODE project. Of these regions, 60% were annotated as intergenic, which implies that they do not overlap with protein-coding genes. Focusing on the intergenic regions, and using stringent analysis of the allele-specific expression data, we detected robust cis-regulatory SNPs in 258 out of 489 informative intergenic regions included in the analysis. The cis-regulatory SNPs that were significantly associated with allele-specific expression of long non-coding RNAs were enriched to enhancer regions marked for active or bivalent, poised chromatin by histone modifications. Out of the lncRNA regions regulated by cis-acting regulatory SNPs, 20% (n = 52) were co-regulated with the closest protein coding gene. We compared the identified cis-regulatory SNPs with those in the catalog of SNPs identified by genome-wide association studies of human diseases and traits. This comparison identified 32 SNPs in loci from genome-wide association studies that displayed a strong association signal with allele-specific expression of non-coding RNAs in monocytes, with p-values ranging from 6.7×10(-7) to 9.5×10(-89). The identified cis-regulatory SNPs are associated with diseases of the immune system, like multiple sclerosis and rheumatoid arthritis.
Conflict of interest statement
Figures
References
-
- Pastinen T, Hudson TJ (2004) Cis-acting regulatory variation in the human genome. Science 306: 647–650. - PubMed
-
- Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, et al. (2009) Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41: 1216–1222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

