Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex
- PMID: 25028510
- PMCID: PMC4148890
- DOI: 10.1074/jbc.M114.566174
Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex
Abstract
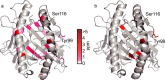
In immune-mediated control of pathogens, human leukocyte antigen (HLA) class I presents various antigenic peptides to CD8(+) T-cells. Long-lived peptide presentation is important for efficient antigen-specific T-cell activation. Presentation time depends on the peptide sequence and the stability of the peptide-HLA complex (pHLA). However, the determinant of peptide-dependent pHLA stability remains elusive. Here, to reveal the pHLA stabilization mechanism, we examined the crystal structures of an HLA class I allomorph in complex with HIV-derived peptides and evaluated site-specific conformational fluctuations using NMR. Although the crystal structures of various pHLAs were almost identical independent of the peptides, fluctuation analyses identified a peptide-dependent minor state that would be more tightly packed toward the peptide. The minor population correlated well with the thermostability and cell surface presentation of pHLA, indicating that this newly identified minor state is important for stabilizing the pHLA and facilitating T-cell recognition.
Keywords: Major Histocompatibility Complex (MHC); Nuclear Magnetic Resonance (NMR); Protein Dynamic; Protein Stability; Thermodynamics.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures










References
-
- Garcia-Lora A., Algarra I., Garrido F. (2003) MHC class I antigens, immune surveillance, and tumor immune escape. J. Cell. Physiol. 195, 346–355 - PubMed
-
- Singer D. S., Mozes E., Kirshner S., Kohn L. D. (1997) Role of MHC class I molecules in autoimmune disease. Crit. Rev. Immunol. 17, 463–468 - PubMed
-
- Goulder P. J., Altfeld M. A., Rosenberg E. S., Nguyen T., Tang Y., Eldridge R. L., Addo M. M., He S., Mukherjee J. S., Phillips M. N., Bunce M., Kalams S. A., Sekaly R. P., Walker B. D., Brander C. (2001) Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193, 181–194 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

