Development and validation of a new high-throughput method to investigate the clonality of HTLV-1-infected cells based on provirus integration sites
- PMID: 25028597
- PMCID: PMC4097847
- DOI: 10.1186/gm568
Development and validation of a new high-throughput method to investigate the clonality of HTLV-1-infected cells based on provirus integration sites
Abstract
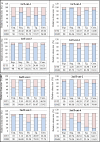
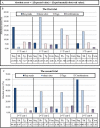
Transformation and clonal proliferation of T-cells infected with human T-cell leukemia virus type-I (HTLV-1) cause adult T-cell leukemia. We took advantage of next-generation sequencing technology to develop and internally validate a new methodology for isolating integration sites and estimating the number of cells in each HTLV-1-infected clone (clone size). Initial analysis was performed with DNA samples from infected individuals. We then used appropriate controls with known integration sites and clonality status to confirm the accuracy of our system, which indeed had the least errors among the currently available techniques. Results suggest potential clinical and biological applications of the new method.
Figures




References
-
- Matsuoka M, Jeang K-T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

