Sequence-based appraisal of the genes encoding neck and carbohydrate recognition domain of conglutinin in blackbuck (Antilope cervicapra) and goat (Capra hircus)
- PMID: 25028649
- PMCID: PMC4083776
- DOI: 10.1155/2014/389150
Sequence-based appraisal of the genes encoding neck and carbohydrate recognition domain of conglutinin in blackbuck (Antilope cervicapra) and goat (Capra hircus)
Abstract
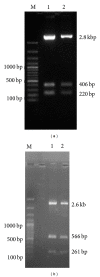
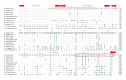
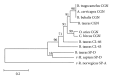
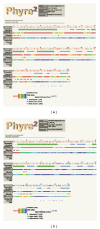
Conglutinin, a collagenous C-type lectin, acts as soluble pattern recognition receptor (PRR) in recognition of pathogens. In the present study, genes encoding neck and carbohydrate recognition domain (NCRD) of conglutinin in goat and blackbuck were amplified, cloned, and sequenced. The obtained 488 bp ORFs encoding NCRD were submitted to NCBI with accession numbers KC505182 and KC505183. Both nucleotide and predicted amino acid sequences were analysed with sequences of other ruminants retrieved from NCBI GenBank using DNAstar and Megalign5.2 software. Sequence analysis revealed maximum similarity of blackbuck sequence with wild ruminants like nilgai and buffalo, whereas goat sequence displayed maximum similarity with sheep sequence at both nucleotide and amino acid level. Phylogenetic analysis further indicated clear divergence of wild ruminants from the domestic ruminants in separate clusters. The predicted secondary structures of NCRD protein in goat and blackbuck using SWISSMODEL ProtParam online software were found to possess 6 beta-sheets and 3 alpha-helices which are identical to the result obtained in case of sheep, cattle, buffalo, and nilgai. However, quaternary structure in goat, sheep, and cattle was found to differ from that of buffalo, nilgai, and blackbuck, suggesting a probable variation in the efficiency of antimicrobial activity among wild and domestic ruminants.
Figures



References
-
- Suzuki Y, Yin Y, Makino M, Kurimura T, Wakamiya N. Cloning and sequencing of a cDNA coding for bovine conglutinin. Biochemical and Biophysical Research Communications. 1993;191(2):335–342. - PubMed
-
- Liou LS, Sastry R, Hartshorn KL, et al. Bovine conglutinin (BC) mRNA expressed in liver: cloning and characterization of the BC cDNA reveals strong homology to surfactant protein-D. Gene. 1994;141(2):277–281. - PubMed
-
- Bordet J, Gay FP. Sur les relations des sensibilisatrices avec l'alexine. Annales de L’Institut Pasteur. 1906;20:467–473.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

