Plasma membrane proteomics of human breast cancer cell lines identifies potential targets for breast cancer diagnosis and treatment
- PMID: 25029196
- PMCID: PMC4100819
- DOI: 10.1371/journal.pone.0102341
Plasma membrane proteomics of human breast cancer cell lines identifies potential targets for breast cancer diagnosis and treatment
Abstract
The use of broad spectrum chemotherapeutic agents to treat breast cancer results in substantial and debilitating side effects, necessitating the development of targeted therapies to limit tumor proliferation and prevent metastasis. In recent years, the list of approved targeted therapies has expanded, and it includes both monoclonal antibodies and small molecule inhibitors that interfere with key proteins involved in the uncontrolled growth and migration of cancer cells. The targeting of plasma membrane proteins has been most successful to date, and this is reflected in the large representation of these proteins as targets of newer therapies. In view of these facts, experiments were designed to investigate the plasma membrane proteome of a variety of human breast cancer cell lines representing hormone-responsive, ErbB2 over-expressing and triple negative cell types, as well as a benign control. Plasma membranes were isolated by using an aqueous two-phase system, and the resulting proteins were subjected to mass spectrometry analysis. Overall, each of the cell lines expressed some unique proteins, and a number of proteins were expressed in multiple cell lines, but in patterns that did not always follow traditional clinical definitions of breast cancer type. From our data, it can be deduced that most cancer cells possess multiple strategies to promote uncontrolled growth, reflected in aberrant expression of tyrosine kinases, cellular adhesion molecules, and structural proteins. Our data set provides a very rich and complex picture of plasma membrane proteins present on breast cancer cells, and the sorting and categorizing of this data provides interesting insights into the biology, classification, and potential treatment of this prevalent and debilitating disease.
Conflict of interest statement
Figures

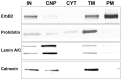

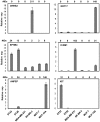

References
-
- Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64: 9–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

