The hypusine-containing translation factor eIF5A
- PMID: 25029904
- PMCID: PMC4183722
- DOI: 10.3109/10409238.2014.939608
The hypusine-containing translation factor eIF5A
Abstract
In addition to the small and large ribosomal subunits, aminoacyl-tRNAs, and an mRNA, cellular protein synthesis is dependent on translation factors. The eukaryotic translation initiation factor 5A (eIF5A) and its bacterial ortholog elongation factor P (EF-P) were initially characterized based on their ability to stimulate methionyl-puromycin (Met-Pmn) synthesis, a model assay for protein synthesis; however, the function of these factors in cellular protein synthesis has been difficult to resolve. Interestingly, a conserved lysine residue in eIF5A is post-translationally modified to hypusine and the corresponding lysine residue in EF-P from at least some bacteria is modified by the addition of a β-lysine moiety. In this review, we provide a summary of recent data that have identified a novel role for the translation factor eIF5A and its hypusine modification in the elongation phase of protein synthesis and more specifically in stimulating the production of proteins containing runs of consecutive proline residues.
Keywords: Deoxyhypusine hydroxylase; deoxyhypusine synthase; elongation factor; polyproline; translation elongation.
Conflict of interest statement
This work was supported by the Intramural Research Program of the National Institutes of Health, NICHD. The authors report no conflict of interest.
Figures
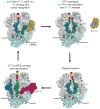

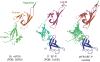

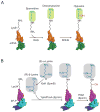
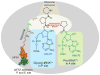
References
-
- Aoki H, Xu J, Emili A, Chosay JG, Golshani A, Ganoza MC. Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 2008;275:671–81. - PubMed
-
- Bartig D, Lemkemeier K, Frank J, Lottspeich F, Klink F. The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur J Biochem. 1992;204:751–8. - PubMed
-
- Bartig D, Schumann H, Klink F. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. Syst Appl Microbiol. 1990;13:112–16.
-
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
