Sequence-specific backbone (1)H, (13)C, and (15)N resonance assignments of human ribonuclease 4
- PMID: 25030111
- PMCID: PMC4764873
- DOI: 10.1007/s12104-014-9570-2
Sequence-specific backbone (1)H, (13)C, and (15)N resonance assignments of human ribonuclease 4
Abstract
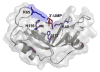
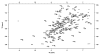
Human ribonuclease 4 (RNase 4) is the most evolutionarily conserved member of the 8 canonical human pancreatic-like RNases, showing more than 90% identity with bovine and porcine homologues. The enzyme displays ribonucleolytic activity with a strong preference for uracil-containing RNA substrates, a feature only shared with human eosinophil derived-neurotoxin (EDN, or RNase 2) and eosinophil cationic protein (ECP, or RNase 3). It is also the shortest member of the human family, with a significantly truncated C-terminal tail. Its unique active-site pocket and high degree of conservation among vertebrates suggest that the enzyme plays a crucial biological function. Here, we report on the (1)H, (13)C and (15)N backbone resonance assignments of RNase 4, providing means to characterize its molecular function at the atomic level by NMR.
Figures

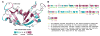
References
-
- Boix E, Blanco JA, Nogues MV, Moussaoui M. Nucleotide binding architecture for secreted cytotoxic endoribonucleases. Biochimie. 2013;95:1087–97. - PubMed
-
- Cranston JW, Perini F, Crisp ER, Hixson CV. Purification and properties of ribonucleases from human urine. Biochim Biophys Acta. 1980;616:239–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

