Reprogramming human endothelial cells to haematopoietic cells requires vascular induction
- PMID: 25030167
- PMCID: PMC4159670
- DOI: 10.1038/nature13547
Reprogramming human endothelial cells to haematopoietic cells requires vascular induction
Abstract
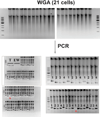
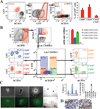
Generating engraftable human haematopoietic cells from autologous tissues is a potential route to new therapies for blood diseases. However, directed differentiation of pluripotent stem cells yields haematopoietic cells that engraft poorly. Here, we have devised a method to phenocopy the vascular-niche microenvironment of haemogenic cells, thereby enabling reprogramming of human endothelial cells into engraftable haematopoietic cells without transition through a pluripotent intermediate. Highly purified non-haemogenic human umbilical vein endothelial cells or adult dermal microvascular endothelial cells were transduced with the transcription factors FOSB, GFI1, RUNX1 and SPI1 (hereafter referred to as FGRS), and then propagated on serum-free instructive vascular niche monolayers to induce outgrowth of haematopoietic colonies containing cells with functional and immunophenotypic features of multipotent progenitor cells (MPPs). These endothelial cells that have been reprogrammed into human MPPs (rEC-hMPPs) acquire colony-forming-cell potential and durably engraft into immune-deficient mice after primary and secondary transplantation, producing long-term rEC-hMPP-derived myeloid (granulocytic/monocytic, erythroid, megakaryocytic) and lymphoid (natural killer and B cell) progenies. Conditional expression of FGRS transgenes, combined with vascular induction, activates endogenous FGRS genes, endowing rEC-hMPPs with a transcriptional and functional profile similar to that of self-renewing MPPs. Our approach underscores the role of inductive cues from the vascular niche in coordinating and sustaining haematopoietic specification and may prove useful for engineering autologous haematopoietic grafts to treat inherited and acquired blood disorders.
Figures



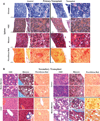
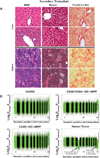
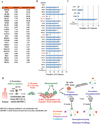


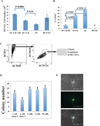




Comment in
-
Stem cells: Reprogramming finds its niche.Nature. 2014 Jul 17;511(7509):301-2. doi: 10.1038/nature13516. Epub 2014 Jul 2. Nature. 2014. PMID: 25030164 Free PMC article.
References
-
- Rafii S, et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood. 2013;121:770–780. - PubMed
-
- Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat Biotechnol. 2013;31:416–418. - PubMed
-
- Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- CA159175/CA/NCI NIH HHS/United States
- CA163167/CA/NCI NIH HHS/United States
- R01 HL115128/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01HL097797/HL/NHLBI NIH HHS/United States
- HL119872/HL/NHLBI NIH HHS/United States
- R01 HL055748/HL/NHLBI NIH HHS/United States
- U54CA163167/CA/NCI NIH HHS/United States
- R01 HL097797/HL/NHLBI NIH HHS/United States
- R01 HL119872/HL/NHLBI NIH HHS/United States
- R21 CA159175/CA/NCI NIH HHS/United States
- U01 HL099997/HL/NHLBI NIH HHS/United States
- HL055748/HL/NHLBI NIH HHS/United States
- U01-HL099997/HL/NHLBI NIH HHS/United States
- U54 CA163167/CA/NCI NIH HHS/United States
- R01 DK095039/DK/NIDDK NIH HHS/United States
- R01HL119872/HL/NHLBI NIH HHS/United States
- R01DK095039/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

