Embryonic stem cell-specific microRNAs contribute to pluripotency by inhibiting regulators of multiple differentiation pathways
- PMID: 25030899
- PMCID: PMC4132708
- DOI: 10.1093/nar/gku544
Embryonic stem cell-specific microRNAs contribute to pluripotency by inhibiting regulators of multiple differentiation pathways
Abstract
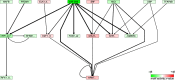
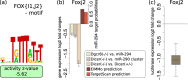
The findings that microRNAs (miRNAs) are essential for early development in many species and that embryonic miRNAs can reprogram somatic cells into induced pluripotent stem cells suggest that these miRNAs act directly on transcriptional and chromatin regulators of pluripotency. To elucidate the transcription regulatory networks immediately downstream of embryonic miRNAs, we extended the motif activity response analysis approach that infers the regulatory impact of both transcription factors (TFs) and miRNAs from genome-wide expression states. Applying this approach to multiple experimental data sets generated from mouse embryonic stem cells (ESCs) that did or did not express miRNAs of the ESC-specific miR-290-295 cluster, we identified multiple TFs that are direct miRNA targets, some of which are known to be active during cell differentiation. Our results provide new insights into the transcription regulatory network downstream of ESC-specific miRNAs, indicating that these miRNAs act on cell cycle and chromatin regulators at several levels and downregulate TFs that are involved in the innate immune response.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. - PubMed
-
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

