Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements
- PMID: 25031611
- PMCID: PMC4100494
- DOI: 10.1186/1756-8935-7-10
Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements
Abstract
Background: Significant efforts have recently been put into the investigation of the spatial organization and the chromatin-interaction networks of genomes. Chromosome conformation capture (3C) technology and its derivatives are important tools used in this effort. However, many of these have limitations, such as being limited to one viewpoint, expensive with moderate to low resolution, and/or requiring a large sequencing effort. Techniques like Hi-C provide a genome-wide analysis. However, it requires massive sequencing effort with considerable costs. Here we describe a new technique termed Targeted Chromatin Capture (T2C), to interrogate large selected regions of the genome. T2C provides an unbiased view of the spatial organization of selected loci at superior resolution (single restriction fragment resolution, from 2 to 6 kbp) at much lower costs than Hi-C due to the lower sequencing effort.
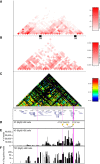
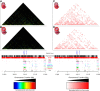
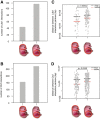
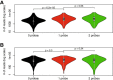
Results: We applied T2C on well-known model regions, the mouse β-globin locus and the human H19/IGF2 locus. In both cases we identified all known chromatin interactions. Furthermore, we compared the human H19/IGF2 locus data obtained from different chromatin conformation capturing methods with T2C data. We observed the same compartmentalization of the locus, but at a much higher resolution (single restriction fragments vs. the common 40 kbp bins) and higher coverage. Moreover, we compared the β-globin locus in two different biological samples (mouse primary erythroid cells and mouse fetal brain), where it is either actively transcribed or not, to identify possible transcriptional dependent interactions. We identified the known interactions in the β-globin locus and the same topological domains in both mouse primary erythroid cells and in mouse fetal brain with the latter having fewer interactions probably due to the inactivity of the locus. Furthermore, we show that interactions due to the important chromatin proteins, Ldb1 and Ctcf, in both tissues can be analyzed easily to reveal their role on transcriptional interactions and genome folding.
Conclusions: T2C is an efficient, easy, and affordable with high (restriction fragment) resolution tool to address both genome compartmentalization and chromatin-interaction networks for specific genomic regions at high resolution for both clinical and non-clinical research.
Keywords: Chromatin conformation capture; Enhancers; Long range interactions; Promoters.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

