Modelling of a genetically diverse evolution of Systemic Mastocytosis with Chronic Myelomonocytic Leukemia (SM-CMML) by Next Generation Sequencing
- PMID: 25032071
- PMCID: PMC4100747
- DOI: 10.1186/2162-3619-3-18
Modelling of a genetically diverse evolution of Systemic Mastocytosis with Chronic Myelomonocytic Leukemia (SM-CMML) by Next Generation Sequencing
Abstract
Background: Systemic mastocytosis (SM) is a heterogenous, clonal mast cell (MC) proliferation, rarely associated with clonal hematologic non-mast cell lineage disease (SM-AHNMD). KIT (D816V) is regarded as driver-mutation in SM-AHNMD.
Methods: DNA isolated from peripheral blood (PB) of an SM-CMML patient was investigated with targeted next generation sequencing. Variants were verified by Sanger sequencing and further characterized in the SM part of the bone marrow trephine (BMT), normal tissue, and FACS sorted PB cell subpopulations.
Findings: Low coverage deep-sequencing (mean 10x) on a GS 454 Junior revealed two as yet unreported SNVs (CBFA2T3 and CLTCL1), both germ-line mutations. High coverage (mean 1674x) targeted re-sequencing on an Ion Proton revealed 177 variants in coding regions. Excluding SNPs, the final list comprised 11 variants. Among these, TET2 (p.Thr1027fs, p.Cys1263Ser) and RUNX1 (p.Asn109Ser) were identified in in the peripheral blood and the SM part of BMT, but not in normal tissue. Furthermore, Sanger sequencing of PB cells revealed similar signal intensities for both TET2 mutations in FACS sorted CD34+ precursor cells and CD16+ granulocytes comparable to signals in the SM part of BMT. In contrast, RUNX1 exhibited a double intensity in CD34+ cells compared to the SM part of BMT and a homozygous variant signal in granulocytes. Both TET2 and RUNX1 mutations were not detectable in B- and T-cells.
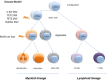
Conclusion: We present a heterozygous triple-mutation pattern (KIT, TET2, RUNX1) in mast cells (SM disease part) with additional LOH of RUNX1 in granulocytes (CMML disease part). These identified mutations allow a more detailed insight into a multistep pathogenesis which suggests a common tumor progenitor in SM-CMML.
Keywords: Chronic myelomonocytic leukemia; Next Generation Sequencing; RUNX1 mutation; SM-CMML; Systemic mastocytosis; TET-2 mutation; c-KIT mutation.
Figures


References
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editor. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2008.
-
- Wang SA, Hutchinson L, Tang G, Chen SS, Miron PM, Huh YO, Jones DM, Bueso-Ramos C, Verstovsek S, Medeiros LJ, Miranda RN. Systemic mastocytosis with associated clonal hematological non-mast cell lineage disease: clinical significance and comparison of chomosomal abnormalities in SM and AHNMD components. Am J Hematol. 2013;88(3):219–224. doi: 10.1002/ajh.23380. - DOI - PMC - PubMed
-
- Pullarkat VA, Bueso-Ramos C, Lai R, Kroft S, Wilson CS, Pullarkat ST, Bu X, Thein M, Lee M, Brynes RK. Systemic mastocytosis with associated clonal hematological non-mast-cell lineage disease: analysis of clinicopathologic features and activating c-kit mutations. Am J Hematol. 2003;73(1):12–17. doi: 10.1002/ajh.10322. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

