Current approaches for predicting a lack of response to anti-EGFR therapy in KRAS wild-type patients
- PMID: 25032217
- PMCID: PMC4086227
- DOI: 10.1155/2014/591867
Current approaches for predicting a lack of response to anti-EGFR therapy in KRAS wild-type patients
Abstract
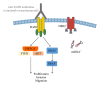
Targeting epidermal growth factor receptor (EGFR) has been one of the most effective colorectal cancer strategies. Anti-EGFR antibodies function by binding to the extracellular domain of EGFR, preventing its activation, and ultimately providing clinical benefit. KRAS mutations in codons 12 and 13 are recognized prognostic and predictive biomarkers that should be analyzed at the clinic prior to the administration of anti-EGFR therapy. However, still an important fraction of KRAS wild-type patients do not respond to the treatment. The identification of additional genetic determinants of primary or secondary resistance to EGFR targeted therapy for further improving the selection of patients is urgent. Herein, we review the latest published literature highlighting the most important genes that may predict resistance to anti-EGFR monoclonal antibodies in colorectal cancer patients. According to the available findings, the evaluation of BRAF, NRAS, PIK3CA, and PTEN status could be the right strategy to select patients who are likely to respond to anti-EGFR therapies. In the future, the combination of those biomarkers will help establish consensus that can be introduced into clinical practice.
Figures


References
-
- Siegel R, Desantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA: A Cancer Journal for Clinicians. 2012;62(4):220–241. - PubMed
-
- Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nature Reviews Cancer. 2009;9(7):489–499. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

