T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection
- PMID: 25032686
- PMCID: PMC4102564
- DOI: 10.1371/journal.ppat.1004251
T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection
Abstract
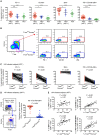
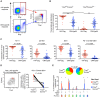
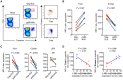
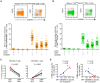
CD8(+) T cell exhaustion represents a major hallmark of chronic HIV infection. Two key transcription factors governing CD8(+) T cell differentiation, T-bet and Eomesodermin (Eomes), have previously been shown in mice to differentially regulate T cell exhaustion in part through direct modulation of PD-1. Here, we examined the relationship between these transcription factors and the expression of several inhibitory receptors (PD-1, CD160, and 2B4), functional characteristics and memory differentiation of CD8(+) T cells in chronic and treated HIV infection. The expression of PD-1, CD160, and 2B4 on total CD8(+) T cells was elevated in chronically infected individuals and highly associated with a T-bet(dim)Eomes(hi) expressional profile. Interestingly, both resting and activated HIV-specific CD8(+) T cells in chronic infection were almost exclusively T-bet(dim)Eomes(hi) cells, while CMV-specific CD8(+) T cells displayed a balanced expression pattern of T-bet and Eomes. The T-bet(dim)Eomes(hi) virus-specific CD8(+) T cells did not show features of terminal differentiation, but rather a transitional memory phenotype with poor polyfunctional (effector) characteristics. The transitional and exhausted phenotype of HIV-specific CD8(+) T cells was longitudinally related to persistent Eomes expression after antiretroviral therapy (ART) initiation. Strikingly, these characteristics remained stable up to 10 years after ART initiation. This study supports the concept that poor human viral-specific CD8(+) T cell functionality is due to an inverse expression balance between T-bet and Eomes, which is not reversed despite long-term viral control through ART. These results aid to explain the inability of HIV-specific CD8(+) T cells to control the viral replication post-ART cessation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, et al. (2005) Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology 6: 1236–1244. - PubMed
-
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, et al. (2003) Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302: 1041–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

