Kernel-PCA data integration with enhanced interpretability
- PMID: 25032747
- PMCID: PMC4101706
- DOI: 10.1186/1752-0509-8-S2-S6
Kernel-PCA data integration with enhanced interpretability
Abstract
Background: Nowadays, combining the different sources of information to improve the biological knowledge available is a challenge in bioinformatics. One of the most powerful methods for integrating heterogeneous data types are kernel-based methods. Kernel-based data integration approaches consist of two basic steps: firstly the right kernel is chosen for each data set; secondly the kernels from the different data sources are combined to give a complete representation of the available data for a given statistical task.
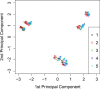
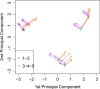
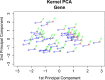
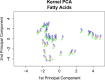
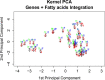
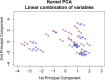
Results: We analyze the integration of data from several sources of information using kernel PCA, from the point of view of reducing dimensionality. Moreover, we improve the interpretability of kernel PCA by adding to the plot the representation of the input variables that belong to any dataset. In particular, for each input variable or linear combination of input variables, we can represent the direction of maximum growth locally, which allows us to identify those samples with higher/lower values of the variables analyzed.
Conclusions: The integration of different datasets and the simultaneous representation of samples and variables together give us a better understanding of biological knowledge.
Figures



References
-
- Gorban AN, Kegl B, Wunsch DC, Zinovyev A. Principal Manifolds for Data Visualization and Dimension Reduction. Springer Publishing Company; 2007.
-
- Pittelkow YE, Wilson SR. Visualisation of Gene Expression Data -the GE-biplot, the Chip-plot and the Gene-plot. Statistical Applications in Genetics and Molecular Biology. 2003. - PubMed
-
- Park M, Lee JW, Lee JB, Song SH. Several biplot methods applied to gene expression data. Journal of Statistical Planning and Inference. 2008;138:500–515. doi: 10.1016/j.jspi.2007.06.019. - DOI
-
- Shawe-Taylor J, Cristianini N. Kernel Methods for Pattern Analysis. Cambridge University Press; 2004.
-
- Scholkopf B, Smola AJ. Learning with Kernels - Support Vector Machines, Regularization, Optimization and Beyond. Cambridge MIT Press; 2002.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

