A comparative pan-genome perspective of niche-adaptable cell-surface protein phenotypes in Lactobacillus rhamnosus
- PMID: 25032833
- PMCID: PMC4102537
- DOI: 10.1371/journal.pone.0102762
A comparative pan-genome perspective of niche-adaptable cell-surface protein phenotypes in Lactobacillus rhamnosus
Abstract
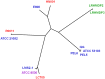
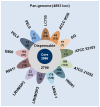
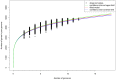
Lactobacillus rhamnosus is a ubiquitously adaptable Gram-positive bacterium and as a typical commensal can be recovered from various microbe-accessible bodily orifices and cavities. Then again, other isolates are food-borne, with some of these having been long associated with naturally fermented cheeses and yogurts. Additionally, because of perceived health benefits to humans and animals, numerous L. rhamnosus strains have been selected for use as so-called probiotics and are often taken in the form of dietary supplements and functional foods. At the genome level, it is anticipated that certain genetic variances will have provided the niche-related phenotypes that augment the flexible adaptiveness of this species, thus enabling its strains to grow and survive in their respective host environments. For this present study, we considered it functionally informative to examine and catalogue the genotype-phenotype variation existing at the cell surface between different L. rhamnosus strains, with the presumption that this might be relatable to habitat preferences and ecological adaptability. Here, we conducted a pan-genomic study involving 13 genomes from L. rhamnosus isolates with various origins. In using a benchmark strain (gut-adapted L. rhamnosus GG) for our pan-genome comparison, we had focused our efforts on a detailed examination and description of gene products for certain functionally relevant surface-exposed proteins, each of which in effect might also play a part in niche adaptability among the other strains. Perhaps most significantly of the surface protein loci we had analyzed, it would appear that the spaCBA operon (known to encode SpaCBA-called pili having a mucoadhesive phenotype) is a genomic rarity and an uncommon occurrence in L. rhamnosus. However, for any of the so-piliated L. rhamnosus strains, they will likely possess an increased niche-specific fitness, which functionally might presumably be manifested by a protracted transient colonization of the gut mucosa or some similar microhabitat.
Conflict of interest statement
Figures

References
-
- Walter J (2005) The microecology of lactobacilli in the gastrointestinal tract. In: Tannock GW, editor. Probiotics and Prebiotics: Scientific Aspects. Norfolk, UK: Caister Academic Press. pp. 51–82.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

