A multiscale approach to modelling drug metabolism by membrane-bound cytochrome P450 enzymes
- PMID: 25033460
- PMCID: PMC4102395
- DOI: 10.1371/journal.pcbi.1003714
A multiscale approach to modelling drug metabolism by membrane-bound cytochrome P450 enzymes
Abstract
Cytochrome P450 enzymes are found in all life forms. P450s play an important role in drug metabolism, and have potential uses as biocatalysts. Human P450s are membrane-bound proteins. However, the interactions between P450s and their membrane environment are not well-understood. To date, all P450 crystal structures have been obtained from engineered proteins, from which the transmembrane helix was absent. A significant number of computational studies have been performed on P450s, but the majority of these have been performed on the solubilised forms of P450s. Here we present a multiscale approach for modelling P450s, spanning from coarse-grained and atomistic molecular dynamics simulations to reaction modelling using hybrid quantum mechanics/molecular mechanics (QM/MM) methods. To our knowledge, this is the first application of such an integrated multiscale approach to modelling of a membrane-bound enzyme. We have applied this protocol to a key human P450 involved in drug metabolism: CYP3A4. A biologically realistic model of CYP3A4, complete with its transmembrane helix and a membrane, has been constructed and characterised. The dynamics of this complex have been studied, and the oxidation of the anticoagulant R-warfarin has been modelled in the active site. Calculations have also been performed on the soluble form of the enzyme in aqueous solution. Important differences are observed between the membrane and solution systems, most notably for the gating residues and channels that control access to the active site. The protocol that we describe here is applicable to other membrane-bound enzymes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



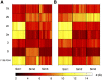


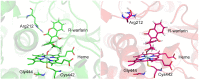

References
-
- Stansfeld PJ, Sansom MSP (2011) Molecular simulation approaches to membrane proteins. Structure 19: 1562–72. - PubMed
-
- Stansfeld PJ, Sansom MSP (2011) From coarse grained to atomistic: A serial multiscale approach to membrane protein simulations. J Chem Theory Comput 7: 1157–1166. - PubMed
-
- van der Kamp MW, Mulholland AJ (2013) Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry 52: 2708–2728. - PubMed
-
- Ortiz de Montellano PR, editor (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd edition. New York: Kluwer Academic/Plenum.
-
- Black SD (1992) Membrane topology of the mammalian P450 cytochromes. FASEB J 6: 680–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/L002558/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/16011/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BEP17032/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- B19456/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H000267/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

