In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner
- PMID: 25034694
- PMCID: PMC4132732
- DOI: 10.1093/nar/gku617
In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner
Abstract
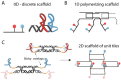
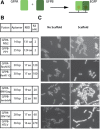
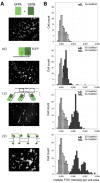
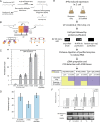
Co-localization of biochemical processes plays a key role in the directional control of metabolic fluxes toward specific products in cells. Here, we employ in vivo scaffolds made of RNA that can bind engineered proteins fused to specific RNA binding domains. This allows proteins to be co-localized on RNA scaffolds inside living Escherichia coli. We assembled a library of eight aptamers and corresponding RNA binding domains fused to partial fragments of fluorescent proteins. New scaffold designs could co-localize split green fluorescent protein fragments to produce activity as measured by cell-based fluorescence. The scaffolds consisted of either single bivalent RNAs or RNAs designed to polymerize in one or two dimensions. The new scaffolds were used to increase metabolic output from a two-enzyme pentadecane production pathway that contains a fatty aldehyde intermediate, as well as three and four enzymes in the succinate production pathway. Pentadecane synthesis depended on the geometry of enzymes on the scaffold, as determined through systematic reorientation of the acyl-ACP reductase fusion by rotation via addition of base pairs to its cognate RNA aptamer. Together, these data suggest that intra-cellular scaffolding of enzymatic reactions may enhance the direct channeling of a variety of substrates.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Agapakis C.M., Boyle P.M., Silver P.A. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 2012;8:527–535. - PubMed
-
- Yeates T.O., Kerfeld C.A., Heinhorst S., Cannon G.C., Shively J.M. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 2008;6:681–691. - PubMed
-
- Srere P.A. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987;56:89–124. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

