Epigenetic targeting of ovarian cancer stem cells
- PMID: 25035395
- PMCID: PMC4155005
- DOI: 10.1158/0008-5472.CAN-14-1022
Epigenetic targeting of ovarian cancer stem cells
Abstract
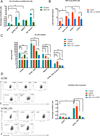
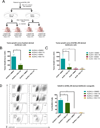
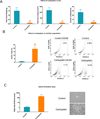
Emerging results indicate that cancer stem-like cells contribute to chemoresistance and poor clinical outcomes in many cancers, including ovarian cancer. As epigenetic regulators play a major role in the control of normal stem cell differentiation, epigenetics may offer a useful arena to develop strategies to target cancer stem-like cells. Epigenetic aberrations, especially DNA methylation, silence tumor-suppressor and differentiation-associated genes that regulate the survival of ovarian cancer stem-like cells (OCSC). In this study, we tested the hypothesis that DNA-hypomethylating agents may be able to reset OCSC toward a differentiated phenotype by evaluating the effects of the new DNA methytransferase inhibitor SGI-110 on OCSC phenotype, as defined by expression of the cancer stem-like marker aldehyde dehydrogenase (ALDH). We demonstrated that ALDH(+) ovarian cancer cells possess multiple stem cell characteristics, were highly chemoresistant, and were enriched in xenografts residual after platinum therapy. Low-dose SGI-110 reduced the stem-like properties of ALDH(+) cells, including their tumor-initiating capacity, resensitized these OCSCs to platinum, and induced reexpression of differentiation-associated genes. Maintenance treatment with SGI-110 after carboplatin inhibited OCSC growth, causing global tumor hypomethylation and decreased tumor progression. Our work offers preclinical evidence that epigenome-targeting strategies have the potential to delay tumor progression by reprogramming residual cancer stem-like cells. Furthermore, the results suggest that SGI-110 might be administered in combination with platinum to prevent the development of recurrent and chemoresistant ovarian cancer.
©2014 American Association for Cancer Research.
Conflict of interest statement
KN and DM received research funding from Astex Pharmaceuticals Inc. and PT is employed by Astex Pharmaceuticals Inc.
Figures



References
-
- Bukowski RM, Ozols RF, Markman M. The management of recurrent ovarian cancer. Seminars in oncology. 2007;34:S1–S15. - PubMed
-
- Liu CM. Cancer of the ovary. The New England journal of medicine. 2005;352:1268–1269. author reply 68–9. - PubMed
-
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. The New England journal of medicine. 2006;355:1253–1261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

