Integrating genomics and proteomics data to predict drug effects using binary linear programming
- PMID: 25036040
- PMCID: PMC4103865
- DOI: 10.1371/journal.pone.0102798
Integrating genomics and proteomics data to predict drug effects using binary linear programming
Abstract
The Library of Integrated Network-Based Cellular Signatures (LINCS) project aims to create a network-based understanding of biology by cataloging changes in gene expression and signal transduction that occur when cells are exposed to a variety of perturbations. It is helpful for understanding cell pathways and facilitating drug discovery. Here, we developed a novel approach to infer cell-specific pathways and identify a compound's effects using gene expression and phosphoproteomics data under treatments with different compounds. Gene expression data were employed to infer potential targets of compounds and create a generic pathway map. Binary linear programming (BLP) was then developed to optimize the generic pathway topology based on the mid-stage signaling response of phosphorylation. To demonstrate effectiveness of this approach, we built a generic pathway map for the MCF7 breast cancer cell line and inferred the cell-specific pathways by BLP. The first group of 11 compounds was utilized to optimize the generic pathways, and then 4 compounds were used to identify effects based on the inferred cell-specific pathways. Cross-validation indicated that the cell-specific pathways reliably predicted a compound's effects. Finally, we applied BLP to re-optimize the cell-specific pathways to predict the effects of 4 compounds (trichostatin A, MS-275, staurosporine, and digoxigenin) according to compound-induced topological alterations. Trichostatin A and MS-275 (both HDAC inhibitors) inhibited the downstream pathway of HDAC1 and caused cell growth arrest via activation of p53 and p21; the effects of digoxigenin were totally opposite. Staurosporine blocked the cell cycle via p53 and p21, but also promoted cell growth via activated HDAC1 and its downstream pathway. Our approach was also applied to the PC3 prostate cancer cell line, and the cross-validation analysis showed very good accuracy in predicting effects of 4 compounds. In summary, our computational model can be used to elucidate potential mechanisms of a compound's efficacy.
Conflict of interest statement
Figures
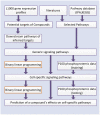

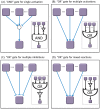

 and
and  , respectively.
, respectively.
References
-
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, et al. (2006) The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–1935. - PubMed
-
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK (2006) Physicochemical modelling of cell signalling pathways. Nature Cell Biology 8: 1195–1203. - PubMed
-
- Kholodenko BN, Demin OV, Moehren G, Hoek JB (1999) Quantification of short term signaling by the epidermal growth factor receptor. Journal of Biological Chemistry 274: 30169–30181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

