Molecular comparisons of full length metapneumovirus (MPV) genomes, including newly determined French AMPV-C and -D isolates, further supports possible subclassification within the MPV Genus
- PMID: 25036224
- PMCID: PMC4103871
- DOI: 10.1371/journal.pone.0102740
Molecular comparisons of full length metapneumovirus (MPV) genomes, including newly determined French AMPV-C and -D isolates, further supports possible subclassification within the MPV Genus
Abstract
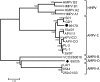
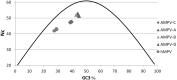
Four avian metapneumovirus (AMPV) subgroups (A-D) have been reported previously based on genetic and antigenic differences. However, until now full length sequences of the only known isolates of European subgroup C and subgroup D viruses (duck and turkey origin, respectively) have been unavailable. These full length sequences were determined and compared with other full length AMPV and human metapneumoviruses (HMPV) sequences reported previously, using phylogenetics, comparisons of nucleic and amino acid sequences and study of codon usage bias. Results confirmed that subgroup C viruses were more closely related to HMPV than they were to the other AMPV subgroups in the study. This was consistent with previous findings using partial genome sequences. Closer relationships between AMPV-A, B and D were also evident throughout the majority of results. Three metapneumovirus "clusters" HMPV, AMPV-C and AMPV-A, B and D were further supported by codon bias and phylogenetics. The data presented here together with those of previous studies describing antigenic relationships also between AMPV-A, B and D and between AMPV-C and HMPV may call for a subclassification of metapneumoviruses similar to that used for avian paramyxoviruses, grouping AMPV-A, B and D as type I metapneumoviruses and AMPV-C and HMPV as type II.
Conflict of interest statement
Figures




Similar articles
-
Comparison of the full-length genome sequence of avian metapneumovirus subtype C with other paramyxoviruses.Virus Res. 2005 Jan;107(1):83-92. doi: 10.1016/j.virusres.2004.07.002. Virus Res. 2005. PMID: 15567037
-
Subtype B avian metapneumovirus resembles subtype A more closely than subtype C or human metapneumovirus with respect to the phosphoprotein, and second matrix and small hydrophobic proteins.Virus Res. 2003 Apr;92(2):171-8. doi: 10.1016/s0168-1702(03)00041-8. Virus Res. 2003. PMID: 12686426
-
Evolutionary dynamics of human and avian metapneumoviruses.J Gen Virol. 2008 Dec;89(Pt 12):2933-2942. doi: 10.1099/vir.0.2008/006957-0. J Gen Virol. 2008. PMID: 19008378
-
Zoonotic Origins of Human Metapneumovirus: A Journey from Birds to Humans.Viruses. 2022 Mar 25;14(4):677. doi: 10.3390/v14040677. Viruses. 2022. PMID: 35458407 Free PMC article. Review.
-
Metapneumoviruses in birds and humans.Virus Res. 2003 Feb;91(2):163-9. doi: 10.1016/s0168-1702(02)00256-3. Virus Res. 2003. PMID: 12573494 Review.
Cited by
-
Trypsin- and low pH-mediated fusogenicity of avian metapneumovirus fusion proteins is determined by residues at positions 100, 101 and 294.Sci Rep. 2015 Oct 26;5:15584. doi: 10.1038/srep15584. Sci Rep. 2015. PMID: 26498473 Free PMC article.
-
Proteome Analysis in a Mammalian Cell line Reveals that PLK2 is Involved in Avian Metapneumovirus Type C (aMPV/C)-Induced Apoptosis.Viruses. 2020 Mar 28;12(4):375. doi: 10.3390/v12040375. Viruses. 2020. PMID: 32231136 Free PMC article.
-
First report of Avian metapneumovirus type B in Iraqi broiler flocks with swollen head syndrome.Vet World. 2022 Jan;15(1):16-21. doi: 10.14202/vetworld.2022.16-21. Epub 2022 Jan 8. Vet World. 2022. PMID: 35369601 Free PMC article.
-
Isolation and characterization of avian metapneumovirus subtypes A and B associated with the 2024 disease outbreaks among poultry in the USA.J Clin Microbiol. 2025 Aug 13;63(8):e0033325. doi: 10.1128/jcm.00333-25. Epub 2025 Jul 10. J Clin Microbiol. 2025. PMID: 40637399 Free PMC article.
-
Evolutionary Trajectories of Avian Avulaviruses and Vaccines Compatibilities in Poultry.Vaccines (Basel). 2022 Nov 3;10(11):1862. doi: 10.3390/vaccines10111862. Vaccines (Basel). 2022. PMID: 36366369 Free PMC article.
References
-
- Buys SB, Du Prees JH (1980) A preliminary report on the isolation of a virus causing sinusitis in turkeys in South Africa and attempts to attenuate the virus. Turkeys 28: 36.
-
- Giraud P, Bennejean G, Guittet M, Toquin D (1986) Turkey rhinotracheitis in France: preliminary investigations on a ciliostatic virus. Vet Rec 119: 606–607. - PubMed
-
- McDougall JS, Cook JK (1986) Turkey rhinotracheitis: preliminary investigations. Vet Rec 118: 206–207. - PubMed
-
- Wilding GP, Baxter-Jones C, Grant M (1986) Ciliostatic agent found in rhinotracheitis. Vet Rec 118: 735. - PubMed
-
- Wyeth PJ, Gough RE, Chettle N, Eddy R (1986) Preliminary observations on a virus associated with turkey rhinotracheitis. Vet Rec 119: 139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

