AtGEN1 and AtSEND1, two paralogs in Arabidopsis, possess holliday junction resolvase activity
- PMID: 25037209
- PMCID: PMC4149707
- DOI: 10.1104/pp.114.237834
AtGEN1 and AtSEND1, two paralogs in Arabidopsis, possess holliday junction resolvase activity
Abstract
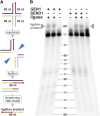
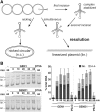
Holliday junctions (HJs) are physical links between homologous DNA molecules that arise as central intermediary structures during homologous recombination and repair in meiotic and somatic cells. It is necessary for these structures to be resolved to ensure correct chromosome segregation and other functions. In eukaryotes, including plants, homologs of a gene called XPG-like endonuclease1 (GEN1) have been identified that process HJs in a manner analogous to the HJ resolvases of phages, archaea, and bacteria. Here, we report that Arabidopsis (Arabidopsis thaliana), a eukaryotic organism, has two functional GEN1 homologs instead of one. Like all known eukaryotic resolvases, AtGEN1 and Arabidopsis single-strand DNA endonuclease1 both belong to class IV of the Rad2/XPG family of nucleases. Their resolvase activity shares the characteristics of the Escherichia coli radiation and UV sensitive C paradigm for resolvases, which involves resolving HJs by symmetrically oriented incisions in two opposing strands. This leads to ligatable products without the need for further processing. The observation that the sequence context influences the cleavage by the enzymes can be interpreted as a hint for the existence of sequence specificity. The two Arabidopsis paralogs differ in their preferred sequences. The precise cleavage positions observed for the resolution of mobile nicked HJs suggest that these cleavage positions are determined by both the substrate structure and the sequence context at the junction point.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures






References
-
- Altona C. (1996) Classification of nucleic acid junctions. J Mol Biol 263: 568–581 - PubMed
-
- Bagherieh-Najjar MB, de Vries OM, Hille J, Dijkwel PP. (2005) Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J 43: 789–798 - PubMed
-
- Bennett RJ, West SC. (1995) Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol 252: 213–226 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

