Dynamic transcriptome landscape of maize embryo and endosperm development
- PMID: 25037214
- PMCID: PMC4149711
- DOI: 10.1104/pp.114.240689
Dynamic transcriptome landscape of maize embryo and endosperm development
Abstract
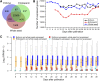
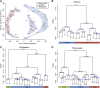
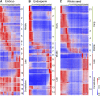
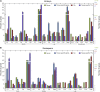
Maize (Zea mays) is an excellent cereal model for research on seed development because of its relatively large size for both embryo and endosperm. Despite the importance of seed in agriculture, the genome-wide transcriptome pattern throughout seed development has not been well characterized. Using high-throughput RNA sequencing, we developed a spatiotemporal transcriptome atlas of B73 maize seed development based on 53 samples from fertilization to maturity for embryo, endosperm, and whole seed tissues. A total of 26,105 genes were found to be involved in programming seed development, including 1,614 transcription factors. Global comparisons of gene expression highlighted the fundamental transcriptomic reprogramming and the phases of development. Coexpression analysis provided further insight into the dynamic reprogramming of the transcriptome by revealing functional transitions during maturation. Combined with the published nonseed high-throughput RNA sequencing data, we identified 91 transcription factors and 1,167 other seed-specific genes, which should help elucidate key mechanisms and regulatory networks that underlie seed development. In addition, correlation of gene expression with the pattern of DNA methylation revealed that hypomethylation of the gene body region should be an important factor for the expressional activation of seed-specific genes, especially for extremely highly expressed genes such as zeins. This study provides a valuable resource for understanding the genetic control of seed development of monocotyledon plants.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Ball SG, Morell MK. (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 - PubMed
-
- Ballicora MA, Iglesias AA, Preiss J. (2004) ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79: 1–24 - PubMed
-
- Bennetzen JL, Hake SC. (2009) Handbook of Maize: Genetics and Genomics. Springer, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

