Eukaryotic origins: How and when was the mitochondrion acquired?
- PMID: 25038049
- PMCID: PMC4292153
- DOI: 10.1101/cshperspect.a015990
Eukaryotic origins: How and when was the mitochondrion acquired?
Abstract
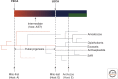
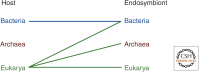
Comparative genomics has revealed that the last eukaryotic common ancestor possessed the hallmark cellular architecture of modern eukaryotes. However, the remarkable success of such analyses has created a dilemma. If key eukaryotic features are ancestral to this group, then establishing the relative timing of their origins becomes difficult. In discussions of eukaryote origins, special significance has been placed on the timing of mitochondrial acquisition. In one view, mitochondrial acquisition was the trigger for eukaryogenesis. Others argue that development of phagocytosis was a prerequisite to acquisition. Results from comparative genomics and molecular phylogeny are often invoked to support one or the other scenario. We show here that the associations between specific cell biological models of eukaryogenesis and evolutionary genomic data are not as strong as many suppose. Disentangling these eliminates many of the arguments that polarize current debate.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
