In-depth characterization of the cerebrospinal fluid (CSF) proteome displayed through the CSF proteome resource (CSF-PR)
- PMID: 25038066
- PMCID: PMC4223498
- DOI: 10.1074/mcp.M114.038554
In-depth characterization of the cerebrospinal fluid (CSF) proteome displayed through the CSF proteome resource (CSF-PR)
Abstract
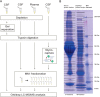
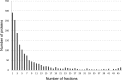
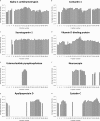
In this study, the human cerebrospinal fluid (CSF) proteome was mapped using three different strategies prior to Orbitrap LC-MS/MS analysis: SDS-PAGE and mixed mode reversed phase-anion exchange for mapping the global CSF proteome, and hydrazide-based glycopeptide capture for mapping glycopeptides. A maximal protein set of 3081 proteins (28,811 peptide sequences) was identified, of which 520 were identified as glycoproteins from the glycopeptide enrichment strategy, including 1121 glycopeptides and their glycosylation sites. To our knowledge, this is the largest number of identified proteins and glycopeptides reported for CSF, including 417 glycosylation sites not previously reported. From parallel plasma samples, we identified 1050 proteins (9739 peptide sequences). An overlap of 877 proteins was found between the two body fluids, whereas 2204 proteins were identified only in CSF and 173 only in plasma. All mapping results are freely available via the new CSF Proteome Resource (http://probe.uib.no/csf-pr), which can be used to navigate the CSF proteome and help guide the selection of signature peptides in targeted quantitative proteomics.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Segal M. B. (1993) Extracellular and cerebrospinal fluids. J. Inherit. Metab. Dis. 16, 617–638 - PubMed
-
- Kroksveen A. C., Opsahl J. A., Aye T. T., Ulvik R. J., Berven F. S. (2011) Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J. Proteomics 74, 371–388 - PubMed
-
- McComb J. G. (1983) Recent research into the nature of cerebrospinal fluid formation and absorption. J. Neurosurg. 59, 369–383 - PubMed
-
- Regeniter A., Kuhle J., Mehling M., Moller H., Wurster U., Freidank H., Siede W. H. (2009) A modern approach to CSF analysis: pathophysiology, clinical application, proof of concept, and laboratory reporting. Clin. Neurol. Neurosurg. 111, 313–318 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

