Comprehensive analysis of loops at protein-protein interfaces for macrocycle design
- PMID: 25038791
- PMCID: PMC4138238
- DOI: 10.1038/nchembio.1580
Comprehensive analysis of loops at protein-protein interfaces for macrocycle design
Abstract
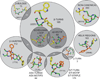
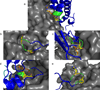
Inhibiting protein-protein interactions (PPIs) with synthetic molecules remains a frontier of chemical biology. Many PPIs have been successfully targeted by mimicking α-helices at interfaces, but most PPIs are mediated by nonhelical, nonstrand peptide loops. We sought to comprehensively identify and analyze these loop-mediated PPIs by writing and implementing LoopFinder, a customizable program that can identify loop-mediated PPIs within all of the protein-protein complexes in the Protein Data Bank. Comprehensive analysis of the entire set of 25,005 interface loops revealed common structural motifs and unique features that distinguish loop-mediated PPIs from other PPIs. 'Hot loops', named in analogy to protein hot spots, were identified as loops with favorable properties for mimicry using synthetic molecules. The hot loops and their binding partners represent new and promising PPIs for the development of macrocycle and constrained peptide inhibitors.
Figures


References
-
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450:1001–1009. - PubMed
-
- Marsault E, Peterson ML. Macrocycles Are Great Cycles: Applications, Opportunities, and Challenges of Synthetic Macrocycles in Drug Discovery. J. Med. Chem. 2011;54:1961–2004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

