From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design
- PMID: 25039866
- PMCID: PMC7163996
- DOI: 10.1111/febs.12936
From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design
Abstract
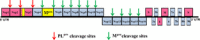
This review focuses on the important contributions that macromolecular crystallography has made over the past 12 years to elucidating structures and mechanisms of the essential proteases of coronaviruses, the main protease (M(pro) ) and the papain-like protease (PL(pro) ). The role of X-ray crystallography in structure-assisted drug discovery against these targets is discussed. Aspects dealt with in this review include the emergence of the SARS coronavirus in 2002-2003 and of the MERS coronavirus 10 years later and the origins of these viruses. The crystal structure of the free SARS coronavirus M(pro) and its dependence on pH is discussed, as are efforts to design inhibitors on the basis of these structures. The mechanism of maturation of the enzyme from the viral polyprotein is still a matter of debate. The crystal structure of the SARS coronavirus PL(pro) and its complex with ubiquitin is also discussed, as is its orthologue from MERS coronavirus. Efforts at predictive structure-based inhibitor development for bat coronavirus M(pro) s to increase the preparedness against zoonotic transmission to man are described as well. The paper closes with a brief discussion of structure-based discovery of antivirals in an academic setting.
Keywords: 3C-like protease; Middle East respiratory syndrome; autoprocessing; bat coronaviruses; high-throughput screening; main protease; papain-like protease; protease maturation; severe acute respiratory syndrome; structure-based inhibitor design.
© 2014 FEBS.
Figures
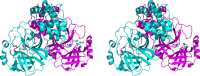


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

