Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis
- PMID: 25042445
- PMCID: PMC4165842
- DOI: 10.1016/j.neuroimage.2014.06.077
Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis
Abstract
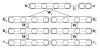
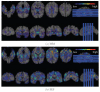
For the last decade, it has been shown that neuroimaging can be a potential tool for the diagnosis of Alzheimer's Disease (AD) and its prodromal stage, Mild Cognitive Impairment (MCI), and also fusion of different modalities can further provide the complementary information to enhance diagnostic accuracy. Here, we focus on the problems of both feature representation and fusion of multimodal information from Magnetic Resonance Imaging (MRI) and Positron Emission Tomography (PET). To our best knowledge, the previous methods in the literature mostly used hand-crafted features such as cortical thickness, gray matter densities from MRI, or voxel intensities from PET, and then combined these multimodal features by simply concatenating into a long vector or transforming into a higher-dimensional kernel space. In this paper, we propose a novel method for a high-level latent and shared feature representation from neuroimaging modalities via deep learning. Specifically, we use Deep Boltzmann Machine (DBM)(2), a deep network with a restricted Boltzmann machine as a building block, to find a latent hierarchical feature representation from a 3D patch, and then devise a systematic method for a joint feature representation from the paired patches of MRI and PET with a multimodal DBM. To validate the effectiveness of the proposed method, we performed experiments on ADNI dataset and compared with the state-of-the-art methods. In three binary classification problems of AD vs. healthy Normal Control (NC), MCI vs. NC, and MCI converter vs. MCI non-converter, we obtained the maximal accuracies of 95.35%, 85.67%, and 74.58%, respectively, outperforming the competing methods. By visual inspection of the trained model, we observed that the proposed method could hierarchically discover the complex latent patterns inherent in both MRI and PET.
Keywords: Alzheimer's Disease; Deep Boltzmann Machine; Mild Cognitive Impairment; Multimodal data fusion; Shared feature representation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

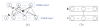
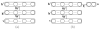


References
-
- Alzheimer’s Association 2012 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia. 2012;8(2):131–168. - PubMed
-
- Baron J, Chtelat G, Desgranges B, Perchey G, Landeau B, de la Sayette V, Eustache F. In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer’s disease. NeuroImage. 2001;14(2):298–309. - PubMed
-
- Belleville S, Clment F, Mellah S, Gilbert B, Fontaine F, Gauthier S. Training-related brain plasticity in subjects at risk of developing Alzheimers disease. Brain. 2011;134(6):1623–1634. - PubMed
-
- Bengio Y. Learning deep architectures for AI. Foundations and Trends in Machine Learning. 2009;2(1):1–127.
-
- Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. In: Schölkopf B, Platt J, Ho man T, editors. Advances in Neural Information Processing Systems 19. MIT Press; Cambridge, MA: 2007. pp. 153–160.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

