The long-term maintenance of a resistance polymorphism through diffuse interactions
- PMID: 25043057
- PMCID: PMC4696508
- DOI: 10.1038/nature13439
The long-term maintenance of a resistance polymorphism through diffuse interactions
Abstract
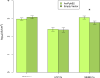
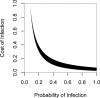
Plant resistance (R) genes are a crucial component in plant defence against pathogens. Although R genes often fail to provide durable resistance in an agricultural context, they frequently persist as long-lived balanced polymorphisms in nature. Standard theory explains the maintenance of such polymorphisms through a balance of the costs and benefits of resistance and virulence in a tightly coevolving host-pathogen pair. However, many plant-pathogen interactions lack such specificity. Whether, and how, balanced polymorphisms are maintained in diffusely interacting species is unknown. Here we identify a naturally interacting R gene and effector pair in Arabidopsis thaliana and its facultative plant pathogen, Pseudomonas syringae. The protein encoded by the R gene RPS5 recognizes an AvrPphB homologue (AvrPphB2) and exhibits a balanced polymorphism that has been maintained for over 2 million years (ref. 3). Consistent with the presence of an ancient balanced polymorphism, the R gene confers a benefit when plants are infected with P. syringae carrying avrPphB2 but also incurs a large cost in the absence of infection. RPS5 alleles are maintained at intermediate frequencies in populations globally, suggesting ubiquitous selection for resistance. However, the presence of P. syringae carrying avrPphB is probably insufficient to explain the RPS5 polymorphism. First, avrPphB homologues occur at very low frequencies in P. syringae populations on A. thaliana. Second, AvrPphB only rarely confers a virulence benefit to P. syringae on A. thaliana. Instead, we find evidence that selection for RPS5 involves multiple non-homologous effectors and multiple pathogen species. These results and an associated model suggest that the R gene polymorphism in A. thaliana may not be maintained through a tightly coupled interaction involving a single coevolved R gene and effector pair. More likely, the stable polymorphism is maintained through complex and diffuse community-wide interactions.
Figures









References
-
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. - PubMed
-
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

