Unraveling recombination rate evolution using ancestral recombination maps
- PMID: 25043668
- PMCID: PMC4314685
- DOI: 10.1002/bies.201400047
Unraveling recombination rate evolution using ancestral recombination maps
Abstract
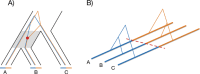
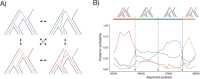
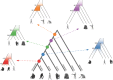
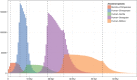
Recombination maps of ancestral species can be constructed from comparative analyses of genomes from closely related species, exemplified by a recently published map of the human-chimpanzee ancestor. Such maps resolve differences in recombination rate between species into changes along individual branches in the speciation tree, and allow identification of associated changes in the genomic sequences. We describe how coalescent hidden Markov models are able to call individual recombination events in ancestral species through inference of incomplete lineage sorting along a genomic alignment. In the great apes, speciation events are sufficiently close in time that a map can be inferred for the ancestral species at each internal branch - allowing evolution of recombination rate to be tracked over evolutionary time scales from speciation event to speciation event. We see this approach as a way of characterizing the evolution of recombination rate and the genomic properties that influence it.
Keywords: evolution; genomics; recombination.
© 2014 The Authors. Bioessays published by WILEY Periodicals, Inc.
Figures
References
-
- Kohli J, Hartsuiker E. Meiosis. eLS. 2001 DOI: 10.1038/npg.els.0001359. - DOI
-
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. - PubMed
-
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. - PubMed
-
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–6. - PubMed
-
- Padhukasahasram B, Rannala B. Meiotic gene-conversion rate and tract length variation in the human genome. Eur J Hum Genet. 2013 in press, DOI: 10.1038/ejhg.2013.30. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

