Functional interplay between protein arginine methyltransferases in Trypanosoma brucei
- PMID: 25044453
- PMCID: PMC4234254
- DOI: 10.1002/mbo3.191
Functional interplay between protein arginine methyltransferases in Trypanosoma brucei
Abstract
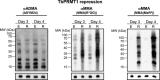
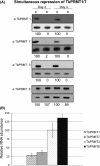
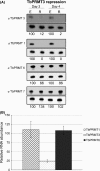
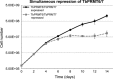
Arginine methylation is a common posttranslational modification that has far-reaching cellular effects. Trypanosoma brucei is an early-branching eukaryote with four characterized protein arginine methyltransferases (PRMTs), one additional putative PRMT, and over 800 arginine methylated proteins, suggesting that arginine methylation has widespread impacts in this organism. While much is known about the activities of individual T. brucei PRMTs (TbPRMTs), little is known regarding how TbPRMTs function together in vivo. In this study, we analyzed single and selected double TbPRMT knockdowns for the impact on expression of TbPRMTs and global methylation status. Repression of TbPRMT1 caused a decrease in asymmetric dimethylarginine and a marked increase in monomethylarginine that was catalyzed by TbPRMT7, suggesting that TbPRMT1 and TbPRMT7 can compete for the same substrate. We also observed an unexpected and strong interdependence between TbPRMT1 and TbPRMT3 protein levels. This finding, together with the observation of similar methyl landscape profiles in TbPRMT1 and TbPRMT3 repressed cells, strongly suggests that these two enzymes form a functional complex. We show that corepression of TbPRMT6/7 synergistically impacts growth of procyclic-form T. brucei. Our findings also implicate the actions of noncanonical, and as yet unidentified, PRMTs in T. brucei. Together, our studies indicate that TbPRMTs display a functional interplay at multiple levels.
Keywords: Arginine methylation; PRMTs; Trypanosomes; posttranslational modifications.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures




References
-
- Anantharaman V, Koonin EV. Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. - PubMed
-
- Anne J. Arginine methylation of SmB is required for Drosophila germ cell development. Development. 2010;137:2819–2828. - PubMed
-
- Bedford MT. Arginine methylation at a glance. J. Cell Sci. 2007;120:4243–4246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

