Mapping the protein interaction landscape for fully functionalized small-molecule probes in human cells
- PMID: 25045785
- PMCID: PMC4120992
- DOI: 10.1021/ja505517t
Mapping the protein interaction landscape for fully functionalized small-molecule probes in human cells
Abstract
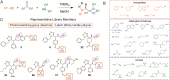
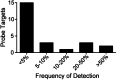
Phenotypic screening provides a means to discover small molecules that perturb cell biological processes. Discerning the proteins and biochemical pathways targeted by screening hits, however, remains technically challenging. We recently described the use of small molecules bearing photoreactive groups and latent affinity handles as fully functionalized probes for integrated phenotypic screening and target identification. The general utility of such probes, or, for that matter, any small-molecule screening library, depends on the scope of their protein interactions in cells, a parameter that remains largely unexplored. Here, we describe the synthesis of an ~60-member fully functionalized probe library, prepared from Ugi-azide condensation reactions to impart structural diversity and introduce diazirine and alkyne functionalities for target capture and enrichment, respectively. In-depth mass spectrometry-based analysis revealed a diverse array of probe targets in human cells, including enzymes, channels, adaptor and scaffolding proteins, and proteins of uncharacterized function. For many of these proteins, ligands have not yet been described. Most of the probe-protein interactions showed well-defined structure-activity relationships across the probe library and were blocked by small-molecule competitors in cells. These findings indicate that fully functionalized small molecules canvas diverse segments of the human proteome and hold promise as pharmacological probes of cell biology.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

