Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules
- PMID: 25046110
- PMCID: PMC4206542
- DOI: 10.4161/auto.29557
Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules
Abstract
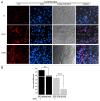
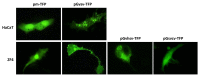
It has not been elucidated whether or not autophagy is induced by rhabdoviral G glycoproteins (G) in vertebrate organisms for which rhabdovirus infection is lethal. Our work provides the first evidence that both mammalian (vesicular stomatitis virus, VSV) and fish (viral hemorrhagic septicemia virus, VHSV, and spring viremia carp virus, SVCV) rhabdoviral Gs induce an autophagic antiviral program in vertebrate cell lines. The transcriptomic profiles obtained from zebrafish genetically immunized with either Gsvcv or Gvhsv suggest that autophagy is induced shortly after immunization and therefore, it may be an important component of the strong antiviral immune responses elicited by these viral proteins. Pepscan mapping of autophagy-inducing linear determinants of Gvhsv and Gvsv showed that peptides located in their fusion domains induce autophagy. Altogether these results suggest that strategies aimed at modulating autophagy could be used for the prevention and treatment of rhabdoviral infections such as rabies, which causes thousands of human deaths every year.
Keywords: LC3; SVCV; VHSV; VSV; antiviral; autophagy; immune response; microarrays; pepscan; rhabdovirus; viral glycoprotein; zebrafish.
Figures

Similar articles
-
VHSV G glycoprotein major determinants implicated in triggering the host type I IFN antiviral response as DNA vaccine molecular adjuvants.Vaccine. 2014 Oct 14;32(45):6012-9. doi: 10.1016/j.vaccine.2014.07.111. Epub 2014 Sep 6. Vaccine. 2014. PMID: 25203447
-
Development of new therapeutical/adjuvant molecules by pepscan mapping of autophagy and IFN inducing determinants of rhabdoviral G proteins.Mol Immunol. 2016 Feb;70:118-24. doi: 10.1016/j.molimm.2015.10.008. Epub 2016 Jan 4. Mol Immunol. 2016. PMID: 26759988 Review.
-
Replication of heterologous glycoprotein-expressing chimeric recombinant snakehead rhabdoviruses (rSHRVs) and viral hemorrhagic septicemia viruses (rVHSVs) at different temperatures.Virus Res. 2021 May;297:198392. doi: 10.1016/j.virusres.2021.198392. Epub 2021 Mar 13. Virus Res. 2021. PMID: 33727091
-
Rainbow Trout Erythrocytes ex vivo Transfection With a DNA Vaccine Encoding VHSV Glycoprotein G Induces an Antiviral Immune Response.Front Immunol. 2018 Oct 29;9:2477. doi: 10.3389/fimmu.2018.02477. eCollection 2018. Front Immunol. 2018. PMID: 30429850 Free PMC article.
-
Spring viremia of carp (SVC).Dis Aquat Organ. 2002 Dec 10;52(3):261-72. doi: 10.3354/dao052261. Dis Aquat Organ. 2002. PMID: 12553453 Review.
Cited by
-
Antigen Presentation and Autophagy in Teleost Adaptive Immunity.Int J Mol Sci. 2022 Apr 28;23(9):4899. doi: 10.3390/ijms23094899. Int J Mol Sci. 2022. PMID: 35563287 Free PMC article. Review.
-
Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers.Cells. 2019 Apr 27;8(5):386. doi: 10.3390/cells8050386. Cells. 2019. PMID: 31035565 Free PMC article.
-
Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish.Vaccines (Basel). 2019 Jul 3;7(3):60. doi: 10.3390/vaccines7030060. Vaccines (Basel). 2019. PMID: 31277329 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
-
Innate response of rainbow trout gill epithelial (RTgill-W1) cell line to ultraviolet-inactivated VHSV and FliC and rhabdovirus infection.Fish Shellfish Immunol Rep. 2021 Dec 10;3:100043. doi: 10.1016/j.fsirep.2021.100043. eCollection 2022 Dec. Fish Shellfish Immunol Rep. 2021. PMID: 36419604 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
