Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosis to LC3-associated pathways involving single- or double-membrane vacuoles
- PMID: 25046114
- PMCID: PMC4206537
- DOI: 10.4161/auto.29411
Role of VAMP3 and VAMP7 in the commitment of Yersinia pseudotuberculosis to LC3-associated pathways involving single- or double-membrane vacuoles
Abstract
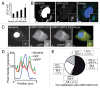
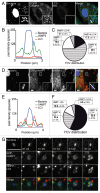
Yersinia pseudotuberculosis can replicate inside macrophages by hijacking autophagy and blocking autophagosome acidification. In bone marrow-derived macrophages, the bacteria are mainly observed inside double-membrane vacuoles positive for LC3, a hallmark of autophagy. Here, we address the question of the membrane traffic during internalization of Yersinia investigating the role of vesicle- associated membrane proteins (VAMPs). First, we show that as in epithelial cells, Yersinia pseudotuberculosis replicates mainly in nonacidic LC3-positive vacuoles. Second, in these cells, we unexpectedly found that VAMP3 localizes preferentially to Yersinia-containing vacuoles (YCVs) with single membranes using correlative light-electron microscopy. Third, we reveal the precise kinetics of VAMP3 and VAMP7 association with YCVs positive for LC3. Fourth, we show that VAMP7 knockdown alters LC3's association with single-and multimembrane-YCVs. Finally, in uninfected epithelial cells stimulated for autophagy, VAMP3 overexpression and knockdown led respectively to a lower and higher number of double-membrane, LC3-positive vesicles. Hence, our results highlight the role that VAMPs play in selection of the pathways leading to generation of ultrastructurally different LC3 compartments and pave the way for determining the full set of docking and fusion proteins involved in Yersinia pseudotuberculosis' intravesicular life cycle.
Keywords: LC3-associated phagosome; SNARE; VAMP3; VAMP7; Yersinia pseudotuberculosis; autophagy; correlative light-electron microscopy.
Figures




Similar articles
-
TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways.Biochim Biophys Acta. 2009 Dec;1793(12):1901-16. doi: 10.1016/j.bbamcr.2009.09.011. Epub 2009 Sep 23. Biochim Biophys Acta. 2009. PMID: 19781582
-
Transport of the major myelin proteolipid protein is directed by VAMP3 and VAMP7.J Neurosci. 2011 Apr 13;31(15):5659-72. doi: 10.1523/JNEUROSCI.6638-10.2011. J Neurosci. 2011. PMID: 21490207 Free PMC article.
-
Isolation and characterization of Salmonella typhimurium and Yersinia pseudotuberculosis-containing phagosomes from infected mouse macrophages: Y. pseudotuberculosis traffics to terminal lysosomes where they are degraded.Eur J Cell Biol. 1998 Sep;77(1):35-47. doi: 10.1016/S0171-9335(98)80100-3. Eur J Cell Biol. 1998. PMID: 9808287
-
Selective autophagy against membranous compartments: Canonical and unconventional purposes and mechanisms.Autophagy. 2014 Mar;10(3):397-407. doi: 10.4161/auto.27244. Epub 2014 Jan 3. Autophagy. 2014. PMID: 24419294 Free PMC article. Review.
-
LC3-associated phagocytosis in microbial pathogenesis.Int J Med Microbiol. 2018 Jan;308(1):228-236. doi: 10.1016/j.ijmm.2017.10.014. Epub 2017 Nov 21. Int J Med Microbiol. 2018. PMID: 29169848 Review.
Cited by
-
Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model.Autophagy. 2019 May;15(5):796-812. doi: 10.1080/15548627.2019.1569297. Epub 2019 Jan 24. Autophagy. 2019. PMID: 30676840 Free PMC article.
-
LAPped in Proof: LC3-Associated Phagocytosis and the Arms Race Against Bacterial Pathogens.Front Cell Infect Microbiol. 2022 Jan 3;11:809121. doi: 10.3389/fcimb.2021.809121. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35047422 Free PMC article. Review.
-
Bacteria Exploit Autophagy For Their Own Benefit.Infect Drug Resist. 2019 Oct 11;12:3205-3215. doi: 10.2147/IDR.S220376. eCollection 2019. Infect Drug Resist. 2019. PMID: 31632106 Free PMC article. Review.
-
LC3-associated phagocytosis: host defense and microbial response.Curr Opin Immunol. 2019 Oct;60:81-90. doi: 10.1016/j.coi.2019.04.012. Epub 2019 Jun 24. Curr Opin Immunol. 2019. PMID: 31247378 Free PMC article. Review.
-
Interactions between Autophagy and Bacterial Toxins: Targets for Therapy?Toxins (Basel). 2015 Aug 4;7(8):2918-58. doi: 10.3390/toxins7082918. Toxins (Basel). 2015. PMID: 26248079 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
