Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers
- PMID: 25047176
- PMCID: PMC4410302
- DOI: 10.1016/j.eururo.2014.06.053
Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers
Abstract
Background: Candidate biomarkers have been identified for clear cell renal cell carcinoma (ccRCC) patients, but most have not been validated.
Objective: To validate published ccRCC prognostic biomarkers in an independent patient cohort and to assess intratumour heterogeneity (ITH) of the most promising markers to guide biomarker optimisation.
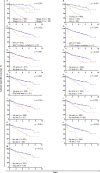
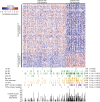
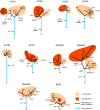
Design, setting, and participants: Cancer-specific survival (CSS) for each of 28 identified genetic or transcriptomic biomarkers was assessed in 350 ccRCC patients. ITH was interrogated in a multiregion biopsy data set of 10 ccRCCs.
Outcome measurements and statistical analysis: Biomarker association with CSS was analysed by univariate and multivariate analyses.
Results and limitations: A total of 17 of 28 biomarkers (TP53 mutations; amplifications of chromosomes 8q, 12, 20q11.21q13.32, and 20 and deletions of 4p, 9p, 9p21.3p24.1, and 22q; low EDNRB and TSPAN7 expression and six gene expression signatures) were validated as predictors of poor CSS in univariate analysis. Tumour stage and the ccB expression signature were the only independent predictors in multivariate analysis. ITH of the ccB signature was identified in 8 of 10 tumours. Several genetic alterations that were significant in univariate analysis were enriched, and chromosomal instability indices were increased in samples expressing the ccB signature. The study may be underpowered to validate low-prevalence biomarkers.
Conclusions: The ccB signature was the only independent prognostic biomarker. Enrichment of multiple poor prognosis genetic alterations in ccB samples indicated that several events may be required to establish this aggressive phenotype, catalysed in some tumours by chromosomal instability. Multiregion assessment may improve the precision of this biomarker.
Patient summary: We evaluated the ability of published biomarkers to predict the survival of patients with clear cell kidney cancer in an independent patient cohort. Only one molecular test adds prognostic information to routine clinical assessments. This marker showed good and poor prognosis results within most individual cancers. Future biomarkers need to consider variation within tumours to improve accuracy.
Keywords: Biomarker; Intratumour heterogeneity; Kidney cancer; Personalised medicine; Prognostic marker.
Copyright © 2014 European Association of Urology. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Motzer R.J., Mazumdar M., Bacik J., Berg W., Amsterdam A., Ferrara J. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J Clin Oncol. 1999;17:2530–2540. - PubMed
-
- Heng D.Y., Xie W., Regan M.M., et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol. 2009;27:5794–5799. - PubMed
-
- Tang P.A., Vickers M.M., Heng D.Y. Clinical and molecular prognostic factors in renal cell carcinoma: what we know so far. Hematol Oncol Clin North Am. 2011;25:871–891. - PubMed
-
- Frank I., Blute M.L., Cheville J.C., Lohse C.M., Weaver A.L., Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. - PubMed
-
- Zisman A., Pantuck A.J., Dorey F., et al. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

