The role of RecQ helicases in non-homologous end-joining
- PMID: 25048400
- PMCID: PMC4244233
- DOI: 10.3109/10409238.2014.942450
The role of RecQ helicases in non-homologous end-joining
Abstract
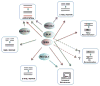
DNA double-strand breaks are highly toxic DNA lesions that cause genomic instability, if not efficiently repaired. RecQ helicases are a family of highly conserved proteins that maintain genomic stability through their important roles in several DNA repair pathways, including DNA double-strand break repair. Double-strand breaks can be repaired by homologous recombination (HR) using sister chromatids as templates to facilitate precise DNA repair, or by an HR-independent mechanism known as non-homologous end-joining (NHEJ) (error-prone). NHEJ is a non-templated DNA repair process, in which DNA termini are directly ligated. Canonical NHEJ requires DNA-PKcs and Ku70/80, while alternative NHEJ pathways are DNA-PKcs and Ku70/80 independent. This review discusses the role of RecQ helicases in NHEJ, alternative (or back-up) NHEJ (B-NHEJ) and microhomology-mediated end-joining (MMEJ) in V(D)J recombination, class switch recombination and telomere maintenance.
Keywords: Alternative end-joining; Ku70/80; RecQ helicases; microhomology-mediated end-joining; non-homologous end-joining; telomere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
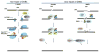
References
-
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. - PubMed
-
- Baynton K, Otterlei M, Bjørås M, et al. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
