Protein painting reveals solvent-excluded drug targets hidden within native protein-protein interfaces
- PMID: 25048602
- PMCID: PMC4109009
- DOI: 10.1038/ncomms5413
Protein painting reveals solvent-excluded drug targets hidden within native protein-protein interfaces
Abstract
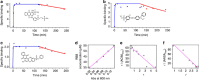
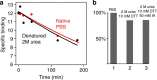
Identifying the contact regions between a protein and its binding partners is essential for creating therapies that block the interaction. Unfortunately, such contact regions are extremely difficult to characterize because they are hidden inside the binding interface. Here we introduce protein painting as a new tool that employs small molecules as molecular paints to tightly coat the surface of protein-protein complexes. The molecular paints, which block trypsin cleavage sites, are excluded from the binding interface. Following mass spectrometry, only peptides hidden in the interface emerge as positive hits, revealing the functional contact regions that are drug targets. We use protein painting to discover contact regions between the three-way interaction of IL1β ligand, the receptor IL1RI and the accessory protein IL1RAcP. We then use this information to create peptides and monoclonal antibodies that block the interaction and abolish IL1β cell signalling. The technology is broadly applicable to discover protein interaction drug targets.
Figures




Comment in
-
Painting a picture of protein interaction.Nat Methods. 2014 Oct;11(10):993. doi: 10.1038/nmeth.3130. Nat Methods. 2014. PMID: 25392882 No abstract available.
References
-
- Nero T. L., Morton C. J., Holien J. K., Wielens J. & Parker M. W. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer 14, 248–262 (2014). - PubMed
-
- Bogan A. A. & Thorn K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998). - PubMed
-
- Arkin M. R. & Wells J. A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3, 301–317 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

