Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers
- PMID: 25048860
- PMCID: PMC4551450
- DOI: 10.1038/nrurol.2014.162
Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers
Abstract
Despite nearly two decades passing since the discovery of gene fusions involving TFE3 or TFEB in sporadic renal cell carcinoma (RCC), the molecular mechanisms underlying the renal-specific tumorigenesis of these genes remain largely unclear. The recently published findings of The Cancer Genome Atlas Network reported that five of the 416 surveyed clear cell RCC tumours (1.2%) harboured SFPQ-TFE3 fusions, providing further evidence for the importance of gene fusions. A total of five TFE3 gene fusions (PRCC-TFE3, ASPSCR1-TFE3, SFPQ-TFE3, NONO-TFE3, and CLTC-TFE3) and one TFEB gene fusion (MALAT1-TFEB) have been identified in RCC tumours and characterized at the mRNA transcript level. A multitude of molecular pathways well-described in carcinogenesis are regulated in part by TFE3 or TFEB proteins, including activation of TGFβ and ETS transcription factors, E-cadherin expression, CD40L-dependent lymphocyte activation, mTORC1 signalling, insulin-dependent metabolism regulation, folliculin signalling, and retinoblastoma-dependent cell cycle arrest. Determining which pathways are most important to RCC oncogenesis will be critical in discovering the most promising therapeutic targets for this disease.
Figures

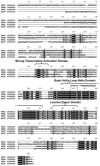



References
-
- Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220:244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

