Diversity-oriented combinatorial biosynthesis of benzenediol lactone scaffolds by subunit shuffling of fungal polyketide synthases
- PMID: 25049383
- PMCID: PMC4151742
- DOI: 10.1073/pnas.1406999111
Diversity-oriented combinatorial biosynthesis of benzenediol lactone scaffolds by subunit shuffling of fungal polyketide synthases
Abstract
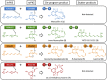
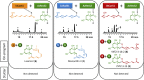
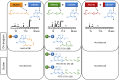
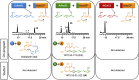
Combinatorial biosynthesis aspires to exploit the promiscuity of microbial anabolic pathways to engineer the synthesis of new chemical entities. Fungal benzenediol lactone (BDL) polyketides are important pharmacophores with wide-ranging bioactivities, including heat shock response and immune system modulatory effects. Their biosynthesis on a pair of sequentially acting iterative polyketide synthases (iPKSs) offers a test case for the modularization of secondary metabolic pathways into "build-couple-pair" combinatorial synthetic schemes. Expression of random pairs of iPKS subunits from four BDL model systems in a yeast heterologous host created a diverse library of BDL congeners, including a polyketide with an unnatural skeleton and heat shock response-inducing activity. Pairwise heterocombinations of the iPKS subunits also helped to illuminate the innate, idiosyncratic programming of these enzymes. Even in combinatorial contexts, these biosynthetic programs remained largely unchanged, so that the iPKSs built their cognate biosynthons, coupled these building blocks into chimeric polyketide intermediates, and catalyzed intramolecular pairing to release macrocycles or α-pyrones. However, some heterocombinations also provoked stuttering, i.e., the relaxation of iPKSs chain length control to assemble larger homologous products. The success of such a plug and play approach to biosynthesize novel chemical diversity bodes well for bioprospecting unnatural polyketides for drug discovery.
Keywords: fungal genetics; secondary metabolites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Fungal polyketide engineering comes of age.Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):12278-9. doi: 10.1073/pnas.1412946111. Epub 2014 Aug 12. Proc Natl Acad Sci U S A. 2014. PMID: 25118274 Free PMC article. No abstract available.
References
-
- Kirschning A, Hahn F. Merging chemical synthesis and biosynthesis: A new chapter in the total synthesis of natural products and natural product libraries. Angew Chem Int Ed Engl. 2012;51(17):4012–4022. - PubMed
-
- Kwon SJ, Mora-Pale M, Lee MY, Dordick JS. Expanding nature’s small molecule diversity via in vitro biosynthetic pathway engineering. Curr Opin Chem Biol. 2012;16(1-2):186–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

