PAPD5-mediated 3' adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease
- PMID: 25049417
- PMCID: PMC4128123
- DOI: 10.1073/pnas.1317751111
PAPD5-mediated 3' adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease
Abstract
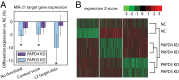
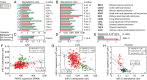
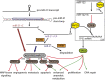
Next-generation sequencing experiments have shown that microRNAs (miRNAs) are expressed in many different isoforms (isomiRs), whose biological relevance is often unclear. We found that mature miR-21, the most widely researched miRNA because of its importance in human disease, is produced in two prevalent isomiR forms that differ by 1 nt at their 3' end, and moreover that the 3' end of miR-21 is posttranscriptionally adenylated by the noncanonical poly(A) polymerase PAPD5. PAPD5 knockdown caused an increase in the miR-21 expression level, suggesting that PAPD5-mediated adenylation of miR-21 leads to its degradation. Exoribonuclease knockdown experiments followed by small-RNA sequencing suggested that PARN degrades miR-21 in the 3'-to-5' direction. In accordance with this model, microarray expression profiling demonstrated that PAPD5 knockdown results in a down-regulation of miR-21 target mRNAs. We found that disruption of the miR-21 adenylation and degradation pathway is a general feature in tumors across a wide range of tissues, as evidenced by data from The Cancer Genome Atlas, as well as in the noncancerous proliferative disease psoriasis. We conclude that PAPD5 and PARN mediate degradation of oncogenic miRNA miR-21 through a tailing and trimming process, and that this pathway is disrupted in cancer and other proliferative diseases.
Keywords: microRNA processing; nucleotidyl transferase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. - PubMed
-
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. - PubMed
-
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

