Next generation sequencing and the future of genetic diagnosis
- PMID: 25052068
- PMCID: PMC4391380
- DOI: 10.1007/s13311-014-0288-8
Next generation sequencing and the future of genetic diagnosis
Abstract
The introduction of next generation sequencing (NGS) has led to an exponential increase of elucidated genetic causes in both extremely rare diseases and common but heterogeneous disorders. It can be applied to the whole or to selected parts of the genome (genome or exome sequencing, gene panels). NGS is not only useful in large extended families with linkage information, but may also be applied to detect de novo mutations or mosaicism in sporadic patients without a prior hypothesis about the mutated gene. Currently, NGS is applied in both research and clinical settings, and there is a rapid transition of research findings to diagnostic applications. These developments may greatly help to minimize the "diagnostic odyssey" for patients as whole-genome analysis can be performed in a few days at reasonable costs compared with gene-by-gene analysis based on Sanger sequencing following diverse clinical tests. Despite the enthusiasm about NGS, one has to keep in mind its limitations, such as a coverage and accuracy of < 100%, resulting in missing variants and false positive findings. In addition, variant interpretation is challenging as there is usually more than one candidate variant found. Therefore, there is an urgent need to define standards for NGS with respect to run quality and variant interpretation, as well as mechanisms of quality control. Further, there are ethical challenges including incidental findings and how to guide unaffected probands seeking direct-to-customer testing. However, taken together, the application of NGS in research and diagnostics provides a tremendous opportunity to better serve our patients.
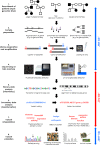
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

