Single cell dissection of early kidney development: multilineage priming
- PMID: 25053437
- PMCID: PMC4197661
- DOI: 10.1242/dev.110601
Single cell dissection of early kidney development: multilineage priming
Abstract
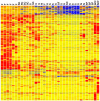
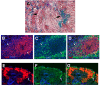
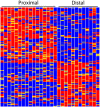
We used a single cell RNA-seq strategy to create an atlas of gene expression patterns in the developing kidney. At several stages of kidney development, histologically uniform populations of cells give rise to multiple distinct lineages. We performed single cell RNA-seq analysis of total mouse kidneys at E11.5 and E12.5, as well as the renal vesicles at P4. We define an early stage of progenitor cell induction driven primarily by gene repression. Surprising stochastic expression of marker genes associated with differentiated cell types was observed in E11.5 progenitors. We provide a global view of the polarized gene expression already present in the renal vesicle, the first epithelial precursor of the nephron. We show that Hox gene read-through transcripts can be spliced to produce intergenic homeobox swaps. We also identify a surprising number of genes with partially degraded noncoding RNA. Perhaps most interesting, at early developmental times single cells often expressed genes related to several developmental pathways. This provides powerful evidence that initial organogenesis involves a process of multilineage priming. This is followed by a combination of gene repression, which turns off the genes associated with most possible lineages, and the activation of increasing numbers of genes driving the chosen developmental direction.
Keywords: Kidney development; Mouse; Multilineage priming; Single cell analysis.
© 2014. Published by The Company of Biologists Ltd.
Figures



References
-
- Boyle S., Misfeldt A., Chandler K. J., Deal K. K., Southard-Smith E. M., Mortlock D. P., Baldwin H. S., de Caestecker M. (2008). Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev. Biol. 313, 234-245 10.1016/j.ydbio.2007.10.014 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

