Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals
- PMID: 25053675
- PMCID: PMC4158756
- DOI: 10.1101/gr.169532.113
Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals
Erratum in
-
Corrigendum: Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals.Genome Res. 2015 Jun;25(6):926. Genome Res. 2015. PMID: 26033277 Free PMC article. No abstract available.
Abstract
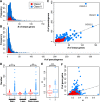
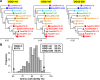
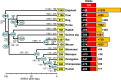
Olfactory receptors (ORs) detect odors in the environment, and OR genes constitute the largest multigene family in mammals. Numbers of OR genes vary greatly among species--reflecting the respective species' lifestyles--and this variation is caused by frequent gene gains and losses during evolution. However, whether the extent of gene gains/losses varies among individual gene lineages and what might generate such variation is unknown. To answer these questions, we used a newly developed phylogeny-based method to classify >10,000 intact OR genes from 13 placental mammal species into 781 orthologous gene groups (OGGs); we then compared the OGGs. Interestingly, African elephants had a surprisingly large repertoire (∼ 2000) of functional OR genes encoded in enlarged gene clusters. Additionally, OR gene lineages that experienced more gene duplication had weaker purifying selection, and Class II OR genes have evolved more dynamically than those in Class I. Some OGGs were highly expanded in a lineage-specific manner, while only three OGGs showed complete one-to-one orthology among the 13 species without any gene gains/losses. These three OGGs also exhibited highly conserved amino acid sequences; therefore, ORs in these OGGs may have physiologically important functions common to every placental mammal. This study provides a basis for inferring OR functions from evolutionary trajectory.
© 2014 Niimura et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Bates LA, Sayialel KN, Njiraini NW, Moss CJ, Poole JH, Byrne RW. 2007. Elephants classify human ethnic groups by odor and garment color. Curr Biol 17: 1938–1942 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
