Single-cell RNA-seq: advances and future challenges
- PMID: 25053837
- PMCID: PMC4132710
- DOI: 10.1093/nar/gku555
Single-cell RNA-seq: advances and future challenges
Abstract
Phenotypically identical cells can dramatically vary with respect to behavior during their lifespan and this variation is reflected in their molecular composition such as the transcriptomic landscape. Single-cell transcriptomics using next-generation transcript sequencing (RNA-seq) is now emerging as a powerful tool to profile cell-to-cell variability on a genomic scale. Its application has already greatly impacted our conceptual understanding of diverse biological processes with broad implications for both basic and clinical research. Different single-cell RNA-seq protocols have been introduced and are reviewed here-each one with its own strengths and current limitations. We further provide an overview of the biological questions single-cell RNA-seq has been used to address, the major findings obtained from such studies, and current challenges and expected future developments in this booming field.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

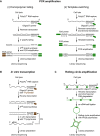

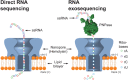
References
-
- Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. - PubMed
-
- Snijder B., Sacher R., Ramo P., Damm E.M., Liberali P., Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. - PubMed
-
- Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., Klein C., Saini M., Bauerle T., Wallwiener M., et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013;31:539–544. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

