Mining TCGA data using Boolean implications
- PMID: 25054200
- PMCID: PMC4108374
- DOI: 10.1371/journal.pone.0102119
Mining TCGA data using Boolean implications
Abstract
Boolean implications (if-then rules) provide a conceptually simple, uniform and highly scalable way to find associations between pairs of random variables. In this paper, we propose to use Boolean implications to find relationships between variables of different data types (mutation, copy number alteration, DNA methylation and gene expression) from the glioblastoma (GBM) and ovarian serous cystadenoma (OV) data sets from The Cancer Genome Atlas (TCGA). We find hundreds of thousands of Boolean implications from these data sets. A direct comparison of the relationships found by Boolean implications and those found by commonly used methods for mining associations show that existing methods would miss relationships found by Boolean implications. Furthermore, many relationships exposed by Boolean implications reflect important aspects of cancer biology. Examples of our findings include cis relationships between copy number alteration, DNA methylation and expression of genes, a new hierarchy of mutations and recurrent copy number alterations, loss-of-heterozygosity of well-known tumor suppressors, and the hypermethylation phenotype associated with IDH1 mutations in GBM. The Boolean implication results used in the paper can be accessed at http://crookneck.stanford.edu/microarray/TCGANetworks/.
Conflict of interest statement
Figures
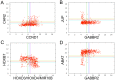

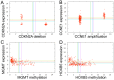
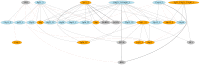
References
-
- van Wieringen W, van de Wiel M (2009) Nonparametric testing for DNA copy number induced differential mrna gene expression. Biometrics 65: 19–29. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

